7O5E
 
 | |
5X2G
 
 | | Crystal structure of Campylobacter jejuni Cas9 in complex with sgRNA and target DNA (AGAAACC PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, Non-target DNA strand, ... | | Authors: | Yamada, M, Watanabe, Y, Hirano, H, Nakane, T, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems
Mol. Cell, 65, 2017
|
|
5X2H
 
 | | Crystal structure of Campylobacter jejuni Cas9 in complex with sgRNA and target DNA (AGAAACA PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, Non-target DNA strand, ... | | Authors: | Yamada, M, Watanabe, Y, Hirano, H, Nakane, T, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems
Mol. Cell, 65, 2017
|
|
1XT8
 
 | | Crystal Structure of Cysteine-Binding Protein from Campylobacter jejuni at 2.0 A Resolution | | Descriptor: | CYSTEINE, GLYCEROL, putative amino-acid transporter periplasmic solute-binding protein | | Authors: | Muller, A, Thomas, G.H, Horler, R, Brannigan, J.A, Blagova, E, Levdikov, V.M, Fogg, M.J, Wilson, K.S, Wilkinson, A.J, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-10-21 | | Release date: | 2005-08-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An ATP-binding cassette-type cysteine transporter in Campylobacter jejuni inferred from the structure of an extracytoplasmic solute receptor protein.
Mol.Microbiol., 57, 2005
|
|
5XBJ
 
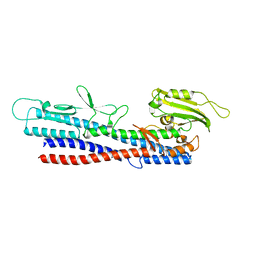 | |
8KFT
 
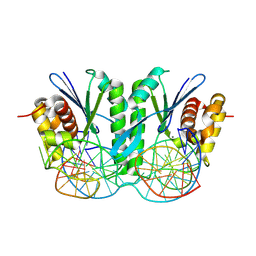 | | Crystal structure of ZmMOC1 in complex with a nicked Holliday junction soaked in Mn2+ for 15 seconds | | Descriptor: | DNA (25-MER), DNA (33-MER), DNA (5'-D(P*CP*AP*CP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Zhang, D, Luo, Z, Lin, Z. | | Deposit date: | 2023-08-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8KFV
 
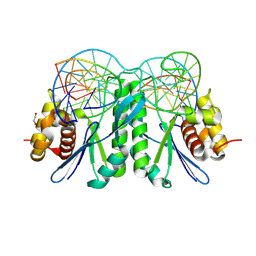 | | Crystal structure of ZmMOC1 K229A in complex with a nicked Holliday junction soaked in Mn2+ for 180 seconds | | Descriptor: | 1,2-ETHANEDIOL, DNA (25-MER), DNA (33-MER), ... | | Authors: | Zhang, D, Luo, Z, Lin, Z. | | Deposit date: | 2023-08-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8KFU
 
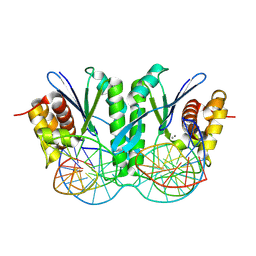 | | Crystal structure of ZmMOC1 in complex with a nicked Holliday junction soaked in Mn2+ for 180 seconds | | Descriptor: | 1,2-ETHANEDIOL, DNA (25-MER), DNA (33-MER), ... | | Authors: | Zhang, D, Luo, Z, Lin, Z. | | Deposit date: | 2023-08-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8KFR
 
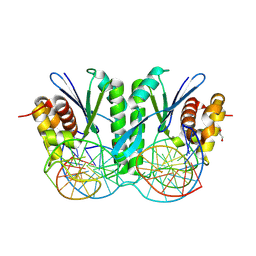 | | Crystal structure of ZmMOC1/nicked Holliday junction/Ca2+ complex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (25-MER), ... | | Authors: | Zhang, D, Luo, Z, Lin, Z. | | Deposit date: | 2023-08-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8KFW
 
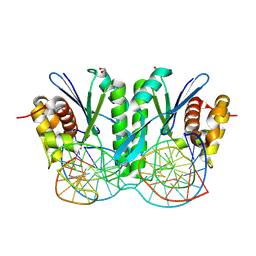 | | Crystal structure of ZmMOC1 K229A in complex with a nicked Holliday junction soaked in Mn2+ for 600 seconds | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (33-MER), ... | | Authors: | Zhang, D, Luo, Z, Lin, Z. | | Deposit date: | 2023-08-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8KFS
 
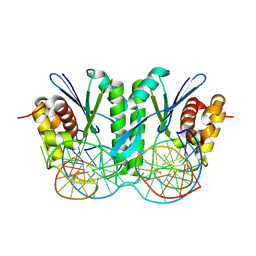 | | Crystal structure of ZmMOC1/nicked Holliday junction complex at ground state | | Descriptor: | DNA (25-MER), DNA (33-MER), DNA (5'-D(P*CP*AP*CP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Zhang, D, Luo, Z, Lin, Z. | | Deposit date: | 2023-08-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
5LQ3
 
 | |
8IEU
 
 | |
8IEV
 
 | |
6CFC
 
 | |
6CF8
 
 | |
8YJX
 
 | |
1D8L
 
 | | E. COLI HOLLIDAY JUNCTION BINDING PROTEIN RUVA NH2 REGION LACKING DOMAIN III | | Descriptor: | PROTEIN (HOLLIDAY JUNCTION DNA HELICASE RUVA) | | Authors: | Nishino, T, Iwasaki, H, Kataoka, M, Ariyoshi, M, Fujita, T, Shinagawa, H, Morikawa, K. | | Deposit date: | 1999-10-25 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Modulation of RuvB function by the mobile domain III of the Holliday junction recognition protein RuvA.
J.Mol.Biol., 298, 2000
|
|
6CF9
 
 | |
3HBN
 
 | | Crystal structure PseG-UDP complex from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, GLYCEROL, UDP-sugar hydrolase, ... | | Authors: | Rangarajan, E.S, Proteau, A, Cygler, M, Matte, A, Sulea, T, Schoenhofen, I.C. | | Deposit date: | 2009-05-04 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional analysis of Campylobacter jejuni PseG: a udp-sugar hydrolase from the pseudaminic acid biosynthetic pathway.
J.Biol.Chem., 284, 2009
|
|
8H2C
 
 | | Crystal structure of the pseudaminic acid synthase PseI from Campylobacter jejuni | | Descriptor: | MANGANESE (II) ION, Pseudaminic acid synthase | | Authors: | Song, W.S, Park, M.A, Ki, D.U, Yoon, S.I. | | Deposit date: | 2022-10-05 | | Release date: | 2022-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural analysis of the pseudaminic acid synthase PseI from Campylobacter jejuni.
Biochem.Biophys.Res.Commun., 635, 2022
|
|
6TZU
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, N84A mutant with pyruvate bound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Lehnert, C, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2019-08-13 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J.Struct.Biol., 209, 2020
|
|
8GR2
 
 | |
8IYG
 
 | | Human neuronal gap junction channel connexin 36 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, DODECYL-BETA-D-MALTOSIDE, Gap junction delta-2 protein | | Authors: | Mao, W.X, Chen, S.S. | | Deposit date: | 2023-04-04 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Assembly mechanisms of the neuronal gap junction channel connexin 36 elucidated by Cryo-EM.
Arch.Biochem.Biophys., 754, 2024
|
|
1RO8
 
 | | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analogue, cytidine-5'-monophosphate | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, alpha-2,3/8-sialyltransferase | | Authors: | Chiu, C.P, Watts, A.G, Lairson, L.L, Gilbert, M, Lim, D, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2003-12-01 | | Release date: | 2004-02-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog.
Nat.Struct.Mol.Biol., 11, 2004
|
|
