5KWA
 
 | |
8KBK
 
 | | Structure of AcrIIA7 complexed with 1',2'-cADPR and cGG | | Descriptor: | (1S,3R,4R,6R,9S,11R,14R,15S,16R,18R)-4-(6-amino-9H-purin-9-yl)-9,11,15,16,18-pentahydroxy-2,5,8,10,12,17-hexaoxa-9lambda~5~,11lambda~5~-diphosphatricyclo[12.2.1.1~3,6~]octadecane-9,11-dione, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Inhibitor of Type II CRISPR-Cas system | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure of AcrIIA7 complexed with 1',2'-cADPR and cGG
To Be Published
|
|
7NFH
 
 | | A heptameric barrel state of a de novo coiled-coil assembly: CC-Type2-(MaId)4. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CC-Type2-(MaId)4 | | Authors: | Rhys, G.G, Dawson, W.M, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2021-02-07 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Differential sensing with arrays of de novo designed peptide assemblies.
Nat Commun, 14, 2023
|
|
7NFN
 
 | | A heptameric barrel state of a de novo coiled-coil assembly: CC-Type2-(LaId)4-L21N-I24N. | | Descriptor: | CC-Type2-(LaId)4-L21N-I24N, HEXANE-1,6-DIOL | | Authors: | Rhys, G.G, Dawson, W.M, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2021-02-07 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Differential sensing with arrays of de novo designed peptide assemblies.
Nat Commun, 14, 2023
|
|
5W2F
 
 | | Crystal Structure of the C-terminal Domain of Human eIF2D at 1.4 A resolution | | Descriptor: | Eukaryotic translation initiation factor 2D, FORMIC ACID | | Authors: | Vaidya, A.T, Lomakin, I.B, Joseph, N.N, Dmitriev, S.E, Steitz, T.A. | | Deposit date: | 2017-06-06 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the C-terminal Domain of Human eIF2D and Its Implications on Eukaryotic Translation Initiation.
J. Mol. Biol., 429, 2017
|
|
5VUL
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 7-(((4-(Dimethylamino)phenethyl)amino)methyl)quinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[({2-[4-(dimethylamino)phenyl]ethyl}amino)methyl]quinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hydrophilic, Potent, and Selective 7-Substituted 2-Aminoquinolines as Improved Human Neuronal Nitric Oxide Synthase Inhibitors.
J. Med. Chem., 60, 2017
|
|
8KCJ
 
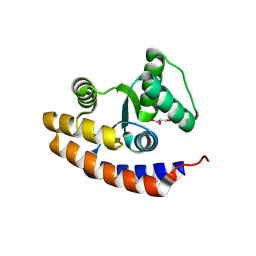 | | De novo design protein -N7 | | Descriptor: | De novo design protein -N7, GLYCEROL | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-07 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | De novo design protein -N7
To Be Published
|
|
7N0E
 
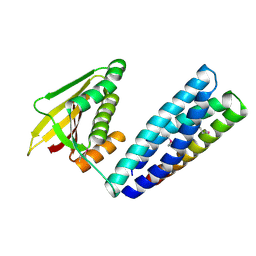 | |
5EEW
 
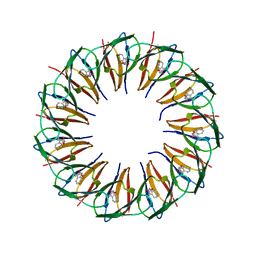 | | RADIATION DAMAGE TO THE TRAP-RNA COMPLEX: DOSE (DWD) 6.45 MGy | | Descriptor: | (GAGUU)10GAG 53-NUCLEOTIDE RNA, TRYPTOPHAN, Transcription attenuation protein MtrB | | Authors: | Bury, C.S, McGeehan, J.E, Garman, E.F, Shevtsov, M.B. | | Deposit date: | 2015-10-23 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | RNA protects a nucleoprotein complex against radiation damage.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7PFN
 
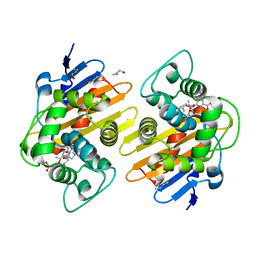 | |
5VUW
 
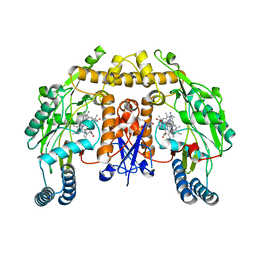 | | Structure of human neuronal nitric oxide synthase heme domain in complex with 7-(((3-(Dimethylamino)benzyl)amino)methyl)quinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[[3-(dimethylamino)phenyl]methylamino]methyl]quinolin-2-amine, Nitric oxide synthase, ... | | Authors: | Huiying, L, Thomas, L.P. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Hydrophilic, Potent, and Selective 7-Substituted 2-Aminoquinolines as Improved Human Neuronal Nitric Oxide Synthase Inhibitors.
J. Med. Chem., 60, 2017
|
|
6M4M
 
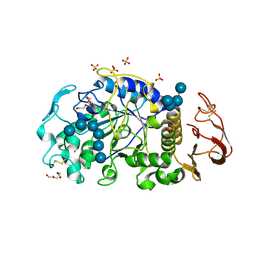 | | X-ray crystal structure of the E249Q mutan of alpha-amylase I and maltohexaose complex from Eisenia fetida | | Descriptor: | Alpha-amylase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hirano, Y, Tsukamoto, K, Ariki, S, Naka, Y, Ueda, M, Tamada, T. | | Deposit date: | 2020-03-07 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray crystallographic structural studies of alpha-amylase I from Eisenia fetida.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7PGO
 
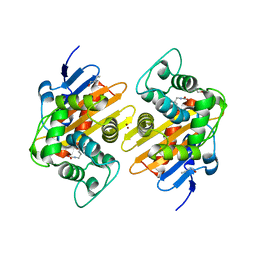 | | Crystal Structure of a Class D Carbapenemase_R250A | | Descriptor: | 1-BUTANOL, BROMIDE ION, Beta-lactamase | | Authors: | Zhou, Q, He, Y, Jin, Y. | | Deposit date: | 2021-08-15 | | Release date: | 2022-08-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An Ion-Pair Induced Intermediate Complex Captured in Class D Carbapenemase Reveals Chloride Ion as a Janus Effector Modulating Activity
Acs Cent.Sci., 2023
|
|
5L72
 
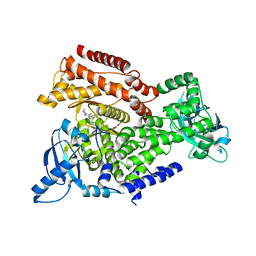 | |
8KB9
 
 | |
6M4V
 
 | | Crystal structure of MBP fused split FKBP in complex with rapamycin | | Descriptor: | GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP1A, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, ... | | Authors: | Kikuchi, M, Wu, D, Inoue, T, Umehara, T. | | Deposit date: | 2020-03-09 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Rational design and implementation of a chemically inducible heterotrimerization system.
Nat.Methods, 17, 2020
|
|
5L7H
 
 | | MCR IN COMPLEX WITH ligand | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 3-methyl-5,8-dioxa-17lambda-thia-4,18-diazatetracyclo[18.2.2.1,.0]pentacosa-1(22),2(6),3,9,11,13(25),20,23-octaene-17,17-dione, Mineralocorticoid receptor, ... | | Authors: | Xue, Y, Aagaard, A, Backstrom, S, Edman, K. | | Deposit date: | 2016-06-03 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-Based Drug Design of Mineralocorticoid Receptor Antagonists to Explore Oxosteroid Receptor Selectivity.
ChemMedChem, 12, 2017
|
|
5VV6
 
 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 7-(((4-(Dimethylamino)phenethyl)amino)methyl)quinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[({2-[4-(dimethylamino)phenyl]ethyl}amino)methyl]quinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-16 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hydrophilic, Potent, and Selective 7-Substituted 2-Aminoquinolines as Improved Human Neuronal Nitric Oxide Synthase Inhibitors.
J. Med. Chem., 60, 2017
|
|
7PIZ
 
 | |
8KC5
 
 | | De novo design protein -T09 | | Descriptor: | De novo design protein -T09 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-06 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | De novo design protein -T09
To Be Published
|
|
5EGK
 
 | |
5ER4
 
 | | Crystal Structure of Calcium-loaded S100B bound to SC0025 | | Descriptor: | 6-methyl-5,6,6~{a},7-tetrahydro-4~{H}-dibenzo[de,g]quinoline-10,11-diol, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Melville, Z.E, Aligholizadeh, E, Fang, L, Alasady, M.J, Pierce, A.D, Wilder, P.T, MacKerell Jr, A.D, Weber, D.J. | | Deposit date: | 2015-11-13 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.813 Å) | | Cite: | Novel protein-inhibitor interactions in site 3 of Ca(2+)-bound S100B as discovered by X-ray crystallography.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5L7O
 
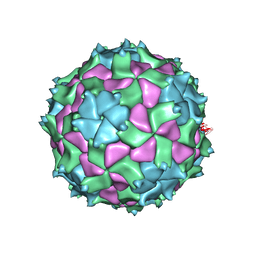 | | X-ray structure of Triatoma virus empty capsid | | Descriptor: | Capsid protein | | Authors: | Sanchez-Eugenia, R. | | Deposit date: | 2016-06-03 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | X-ray structure of Triatoma virus empty capsid: insights into the mechanism of uncoating and RNA release in dicistroviruses.
J.Gen.Virol., 97, 2016
|
|
8C7U
 
 | |
8C7O
 
 | |
