1KBR
 
 | |
6N3I
 
 | |
7CQQ
 
 | | GmaS in complex with AMPPNP and MetSox | | Descriptor: | (2S)-2-AMINO-4-(METHYLSULFONIMIDOYL)BUTANOIC ACID, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQN
 
 | | GmaS in complex with AMPPCP | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
6D2T
 
 | | HLA-B*57:01 presenting LALLTGVRW | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-57 alpha chain, ... | | Authors: | Vivian, J.P, Rossjohn, J. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands.
Sci Immunol, 3, 2018
|
|
4YUU
 
 | | Crystal structure of oxygen-evolving photosystem II from a red alga | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ago, H, Shen, J.-R. | | Deposit date: | 2015-03-19 | | Release date: | 2016-01-20 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (2.7700038 Å) | | Cite: | Novel Features of Eukaryotic Photosystem II Revealed by Its Crystal Structure Analysis from a Red Alga
J.Biol.Chem., 291, 2016
|
|
7CQU
 
 | | GmaS/ADP/MetSox-P complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
3DRQ
 
 | | Crystal structure of the HIV-1 broadly neutralizing antibody 2F5 in complex with the gp41 FP-MPER Hyb3K construct 514GIGALFLGFLGAAGS528KK-Ahx-655KNEQELLELDKWASLWN671 soaked in PEG/2-propanol solution | | Descriptor: | 2F5 Fab' heavy chain, 2F5 Fab' light chain, GLYCEROL, ... | | Authors: | Bryson, S, Julien, J.P, Pai, E.F. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site.
J.Mol.Biol., 384, 2008
|
|
5JNE
 
 | | E2-SUMO-Siz1 E3-SUMO-PCNA complex | | Descriptor: | E3 SUMO-protein ligase SIZ1,Ubiquitin-like protein SMT3, GLYCEROL, Proliferating cell nuclear antigen, ... | | Authors: | Lima, C.D, Streich Jr, F.C. | | Deposit date: | 2016-04-29 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Capturing a substrate in an activated RING E3/E2-SUMO complex.
Nature, 536, 2016
|
|
7CQX
 
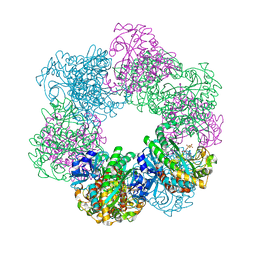 | | GmaS/ADP complex-Conformation 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
4ZHQ
 
 | | Crystal structure of Tubulin-Stathmin-TTL-MMAE Complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Zhang, R. | | Deposit date: | 2015-04-26 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insights into the Pharmacophore of Vinca Domain Inhibitors of Microtubules
Mol.Pharmacol., 89, 2016
|
|
4RZT
 
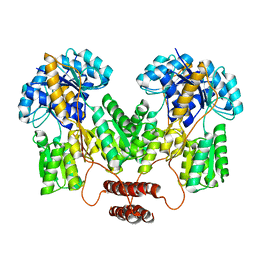 | | Lac repressor engineered to bind sucralose, sucralose-bound tetramer | | Descriptor: | 4-chloro-4-deoxy-alpha-D-galactopyranose-(1-2)-1,6-dichloro-1,6-dideoxy-beta-D-fructofuranose, Lac repressor | | Authors: | Arbing, M.A, Cascio, D, Sawaya, M.R, Kosuri, S, Church, G.M. | | Deposit date: | 2014-12-24 | | Release date: | 2015-12-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Engineering an allosteric transcription factor to respond to new ligands.
Nat.Methods, 13, 2016
|
|
4ZI7
 
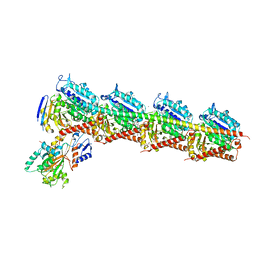 | | CRYSTAL STRUCTURE OF TUBULIN-STATHMIN-TTL-HTI286 COMPLEX | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Zhang, R. | | Deposit date: | 2015-04-27 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural Insights into the Pharmacophore of Vinca Domain Inhibitors of Microtubules
Mol.Pharmacol., 89, 2016
|
|
6D29
 
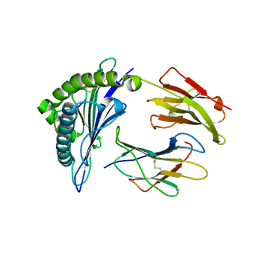 | | HLA-B*57:01 presenting TSMSFVPRPW | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-57 alpha chain, ... | | Authors: | Vivian, J.P, Rossjohn, J. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands.
Sci Immunol, 3, 2018
|
|
2I0C
 
 | |
2PTT
 
 | |
3T6P
 
 | | IAP antagonist-induced conformational change in cIAP1 promotes E3 ligase activation via dimerization | | Descriptor: | Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Dueber, E.C, Schoeffler, A.J, Lingel, A, Elliott, M, Fedorova, A.V, Giannetti, A.M, Zobel, K, Maurer, B, Varfolomeev, E, Wu, P, Wallweber, H, Hymowitz, S, Deshayes, K, Vucic, D, Fairbrother, W.J. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Antagonists induce a conformational change in cIAP1 that promotes autoubiquitination.
Science, 334, 2011
|
|
2EXL
 
 | | GRP94 N-terminal Domain bound to geldanamycin | | Descriptor: | Endoplasmin, GELDANAMYCIN, PENTAETHYLENE GLYCOL, ... | | Authors: | Reardon, P.N, Immormino, R.M, Gewirth, D.T. | | Deposit date: | 2005-11-08 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Different poses for ligand and chaperone in inhibitor-bound Hsp90 and GRP94: implications for paralog-specific drug design.
J.Mol.Biol., 388, 2009
|
|
4J6G
 
 | | CRYSTAL STRUCTURE OF LIGHT AND DcR3 COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Liu, W, Zhan, C, Bonanno, J.B, Bhosle, R.C, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-11 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
3OKM
 
 | | Crystal structure of unliganded S25-39 | | Descriptor: | S25-39 Fab (IgG1k) heavy chain, S25-39 Fab (IgG1k) light chain | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2010-08-25 | | Release date: | 2011-04-06 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Common NH53K Mutation in the Combining Site of Antibodies Raised against Chlamydial LPS Glycoconjugates Significantly Increases Avidity.
Biochemistry, 50, 2011
|
|
2PTV
 
 | | Structure of NK cell receptor ligand CD48 | | Descriptor: | CD48 antigen | | Authors: | Deng, L, Velikovsky, C.A, Mariuzza, R.A. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of natural killer receptor 2B4 bound to CD48 reveals basis for heterophilic recognition in signaling lymphocyte activation molecule family.
Immunity, 27, 2007
|
|
2HDP
 
 | | Solution Structure of Hdm2 RING Finger Domain | | Descriptor: | Ubiquitin-protein ligase E3 Mdm2, ZINC ION | | Authors: | Kostic, M, Matt, T, Yamout-Martinez, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2006-06-20 | | Release date: | 2006-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Hdm2 C2H2C4 RING, a domain critical for ubiquitination of p53.
J.Mol.Biol., 363, 2006
|
|
8XHK
 
 | | Crystal structure of alpha-Oxoamine Synthase Alb29 with PLP cofactor | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Xu, M.J, Zhang, D.K. | | Deposit date: | 2023-12-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural and mechanistic investigations on CC bond forming alpha-oxoamine synthase allowing L-glutamate as substrate.
Int.J.Biol.Macromol., 268, 2024
|
|
9F41
 
 | | Crystal structure of the NTD domain from S. cerevisia Niemann-Pick type C protein NCR1 with cholesterol bound | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHOLESTEROL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nel, L, Olesen, E, Frain, K.M, Pedersen, B.P. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural and biochemical analysis of ligand binding in yeast Niemann-Pick type C 1-related protein
To Be Published
|
|
8VUG
 
 | | Crystal Structure of human Tryptophan 2,3-dioxygenase in complex with PPN1 inhibitor | | Descriptor: | (5P)-5-(1H-indol-3-yl)-1-[(piperidin-4-yl)methyl]-1H-benzotriazole, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase, ... | | Authors: | Geeraerts, Z, Yeh, S. | | Deposit date: | 2024-01-29 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insights into Protein-Inhibitor Interactions in Human Tryptophan Dioxygenase.
J.Med.Chem., 67, 2024
|
|
