8RZH
 
 | | ZgGH129 from Zobellia galactanivorans in complex with the inhibitor AD-DGJ (3,6-anhydro-D-1-deoxygalactonojirimycin). | | Descriptor: | (1~{R},4~{S},5~{R},8~{S})-6-oxa-2-azabicyclo[3.2.1]octane-4,8-diol, 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE, ... | | Authors: | Roret, T, Czjzek, M, Ficko-Blean, E. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
7PGN
 
 | | HHP-C in complex with glycosaminoglycan mimic SOS | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Griffiths, S.C, Schwab, R.A, El Omari, K, Bishop, B, Iverson, E.J, Malinuskas, T, Dubey, R, Qian, M, Covey, D.F, Gilbert, R.J.C, Rohatgi, R, Siebold, C. | | Deposit date: | 2021-08-14 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hedgehog-Interacting Protein is a multimodal antagonist of Hedgehog signalling.
Nat Commun, 12, 2021
|
|
5TW0
 
 | |
8RQG
 
 | |
6N0Z
 
 | |
8OFP
 
 | |
7NNF
 
 | | Crystal structure of Mycobacterium tuberculosis ArgF in apo form. | | Descriptor: | Ornithine carbamoyltransferase, PHOSPHATE ION | | Authors: | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLP
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with L-canavanine | | Descriptor: | Acetylglutamate kinase, L-CANAVANINE, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.213 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
8OFU
 
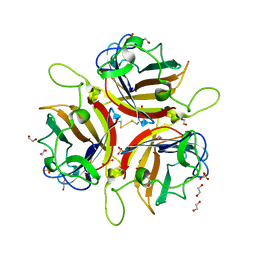 | | Human adenovirus type 53 fiber-knob protein | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Fiber protein, ... | | Authors: | Rizkallah, P.J, Parker, A.L, Mundy, R.M, Baker, A.T. | | Deposit date: | 2023-03-16 | | Release date: | 2023-09-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Broad sialic acid usage amongst species D human adenovirus.
Npj Viruses, 1, 2023
|
|
7ZAL
 
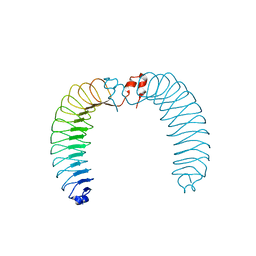 | |
5UGF
 
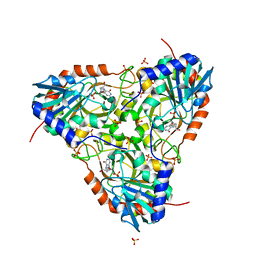 | | Crystal structure of human purine nucleoside phosphorylase (F159Y) mutant complexed with DADMe-ImmG and phosphate | | Descriptor: | 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Harijan, R.K, Cameron, S.A, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-01-08 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic-site design for inverse heavy-enzyme isotope effects in human purine nucleoside phosphorylase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7VU4
 
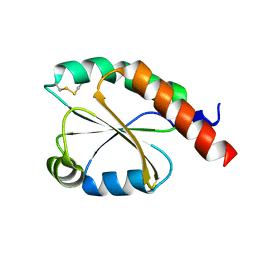 | | de novo design based on 1r26 | | Descriptor: | de novo design protein | | Authors: | Zhang, L. | | Deposit date: | 2021-11-01 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rotamer-free protein sequence design based on deep learning and self-consistency.
Nat Comput Sci, 2023
|
|
7NLX
 
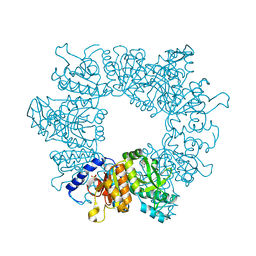 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 7-(trifluoromethyl)quinolin-4-ol. | | Descriptor: | 7-(trifluoromethyl)quinolin-4-ol, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.234 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7VIP
 
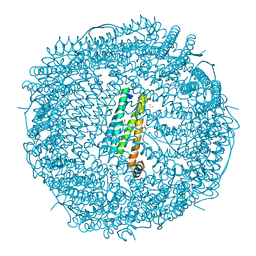 | | Crystal structure of Au(10EQ)-apo-R168H/L169C-rHLFr | | Descriptor: | CADMIUM ION, Ferritin light chain, GLYCEROL, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
7YY3
 
 | |
7NNV
 
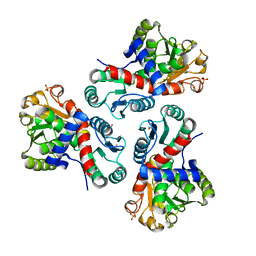 | | Crystal structure of Mycobacterium tuberculosis ArgF in complex with carbamoyl phosphate. | | Descriptor: | 1,2-ETHANEDIOL, Ornithine carbamoyltransferase, PHOSPHATE ION, ... | | Authors: | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | Deposit date: | 2021-02-25 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
8OME
 
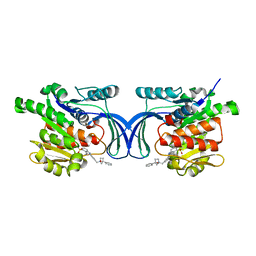 | | Crystal structure of hKHK-A in complex with compound-4 | | Descriptor: | Ketohexokinase, compound | | Authors: | Ebenhoch, R, Pautsch, A. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human and mouse ketohexokinase provide a structural basis for species- and isoform-selective inhibitor design.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5TY1
 
 | | Identification of a New Zinc Binding Chemotype by Fragment Screening | | Descriptor: | (5R)-5-(2,4-dimethoxyphenyl)-1,3-oxazolidine-2,4-dione, Carbonic anhydrase 2, SODIUM ION, ... | | Authors: | Peat, T.S. | | Deposit date: | 2016-11-18 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
8RZ0
 
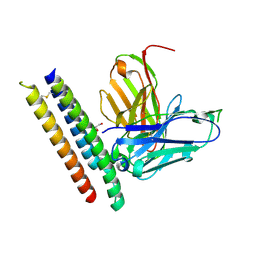 | |
5TY4
 
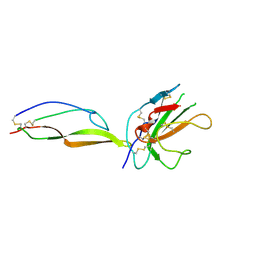 | | MicroED structure of a complex between monomeric TGF-b and its receptor, TbRII, at 2.9 A resolution | | Descriptor: | TGF-beta receptor type-2, mmTGF-b2-7m | | Authors: | Weiss, S.C, de la Cruz, M.J, Hattne, J, Shi, D, Reyes, F.E, Callero, G, Gonen, T. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.9 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
7VTY
 
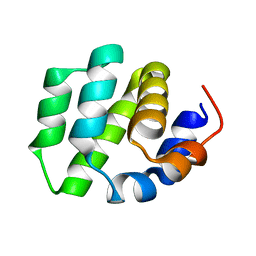 | | de novo designed protein | | Descriptor: | de novo designed protein | | Authors: | Zhang, L. | | Deposit date: | 2021-10-31 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rotamer-free protein sequence design based on deep learning and self-consistency.
Nat Comput Sci, 2023
|
|
8U57
 
 | | PPARg LBD in complex with perfluorooctanoic acid (PFOA) | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, Peroxisome proliferator-activated receptor gamma, pentadecafluorooctanoic acid | | Authors: | Pederick, J.L, Frkic, R.L, McDougal, D.P, Bruning, J.B. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural basis for the activation of peroxisome proliferator-activated receptor gamma (PPAR gamma ) by perfluorooctanoic acid (PFOA).
Chemosphere, 354, 2024
|
|
7NLT
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 4-(4-methylpiperazin-1-yl)benzoic acid | | Descriptor: | 4-(4-methylpiperazin-1-yl)benzoic acid, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.225 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7VTA
 
 | |
5TY9
 
 | | Identification of a New Zinc Binding Chemotype by Fragment Screening | | Descriptor: | (5R)-5-(2,4-dimethoxyphenyl)-1,3-oxazolidine-2,4-dione, Carbonic anhydrase 2, ZINC ION | | Authors: | Peat, T.S, Poulsen, S.A, Ren, B, Dolezal, O, Woods, L.A, Mujumdar, P, Chrysanthopoulos, P.K. | | Deposit date: | 2016-11-18 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
