8BES
 
 | |
1GXZ
 
 | |
5E6A
 
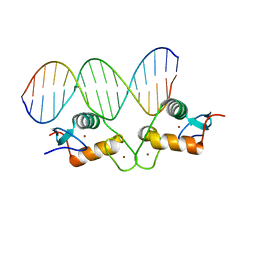 | | Glucocorticoid receptor DNA binding domain - PLAU NF-kB response element complex | | Descriptor: | DNA (5'-D(*AP*TP*CP*AP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*TP*GP*AP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Rye, E.A, Herbst, A.G, Ortlund, E.A. | | Deposit date: | 2015-10-09 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Cryptic glucocorticoid receptor-binding sites pervade genomic NF-kappa B response elements.
Nat Commun, 9, 2018
|
|
6FQP
 
 | |
6TTJ
 
 | | Neutral invertase 2 from Arabidopsis thaliana | | Descriptor: | Alkaline/neutral invertase CINV1 | | Authors: | Tsirkone, V.G, Osipov, E.M, Beelen, S, Strelkov, S.V. | | Deposit date: | 2019-12-27 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.392 Å) | | Cite: | Crystal structure of Arabidopsis thaliana neutral invertase 2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
8BEQ
 
 | | Structure of fructofuranosidase from Rhodotorula dairenensis | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2022-10-21 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Insights into the Structure of the Highly Glycosylated Ffase from Rhodotorula dairenensis Enhance Its Biotechnological Potential.
Int J Mol Sci, 23, 2022
|
|
5WIK
 
 | | JAK2 Pseudokinase in complex with BI-D1870 | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-5,7-dimethyl-8-(3-methylbutyl)-7,8-dihydropteridin-6(5H)-one, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
5WIM
 
 | | JAK2 Pseudokinase in complex with AT9283 | | Descriptor: | 1-cyclopropyl-3-{3-[5-(morpholin-4-ylmethyl)-1H-benzimidazol-2-yl]-1H-pyrazol-4-yl}urea, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
8BEU
 
 | |
6WL3
 
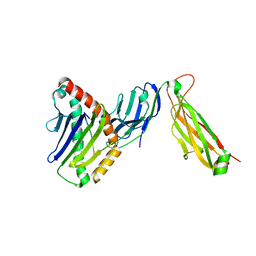 | | preTCRbeta-pMHC complex crystal structure | | Descriptor: | ARG-GLY-TYR-LEU-TYR-GLN-GLY-LEU, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | Authors: | Li, X, Mallis, R.J, Mizsei, R, Tan, K, Reinherz, E.L, Wang, J. | | Deposit date: | 2020-04-18 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Pre-T cell receptors topologically sample self-ligands during thymocyte beta-selection.
Science, 371, 2021
|
|
5WIJ
 
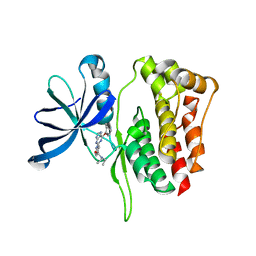 | | JAK2 Pseudokinase in complex with NU6140 | | Descriptor: | 4-{[6-(cyclohexylmethoxy)-7H-purin-2-yl]amino}-N,N-diethylbenzamide, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
1RQF
 
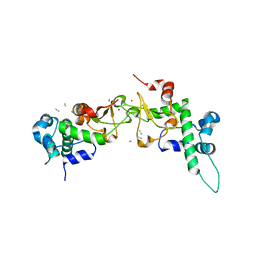 | | Structure of CK2 beta subunit crystallized in the presence of a p21WAF1 peptide | | Descriptor: | Casein kinase II beta chain, Disordered segment of Cyclin-dependent kinase inhibitor 1, UNKNOWN, ... | | Authors: | Bertrand, L, Sayed, M.F, Pei, X.-Y, Parisini, E, Dhanaraj, V, Bolanos-Garcia, V.M, Allende, J.E, Blundell, T.L. | | Deposit date: | 2003-12-05 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of the regulatory subunit of CK2 in the presence of a p21WAF1 peptide demonstrates flexibility of the acidic loop.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6NSZ
 
 | | X-ray reduced Catalase 3 from N.Crassa (0.526 MGy) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Catalase-3, ... | | Authors: | Zarate-Romero, A, Rudino-Pinera, E, Stojanoff, V. | | Deposit date: | 2019-01-27 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray driven reduction of Cpd I of Catalase-3 from N. crassa reveals differential sensitivity of active sites and formation of ferrous state.
Arch.Biochem.Biophys., 666, 2019
|
|
3OZM
 
 | | Crystal structure of enolase superfamily member from Bordetella bronchiseptica complexed with Mg, m-Xylarate and L-Lyxarate | | Descriptor: | D-xylaric acid, GLYCEROL, L-arabinaric acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wichelecki, D, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-09-25 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of enolase superfamily member from Bordetella bronchiseptica complexed with Mg, m-Xylarate and L-Lyxarate
TO BE PUBLISHED
|
|
5EA0
 
 | | Structure of the antibody 7968 with human complement factor H-derived peptide | | Descriptor: | Complement factor H-related protein 2, Heavy chain of antibody 7968 Fab fragment, Light chain of antibody 7968 Fab fragment | | Authors: | Bushey, R.T, Moody, M.A, Nicely, N.I, Alam, S.M, Haynes, B.F, Winkler, M.T, Gottlin, E.B, Campa, M.J, Liao, H.-X, Patz Jr, E.F. | | Deposit date: | 2015-10-15 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Therapeutic Antibody for Cancer, Derived from Single Human B Cells.
Cell Rep, 15, 2016
|
|
6X4T
 
 | | Crystal structure of ICOS-L in complex with Prezalumab and VNAR domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Rujas, E, Sicard, T, Julien, J.P. | | Deposit date: | 2020-05-23 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural characterization of the ICOS/ICOS-L immune complex reveals high molecular mimicry by therapeutic antibodies.
Nat Commun, 11, 2020
|
|
4K8N
 
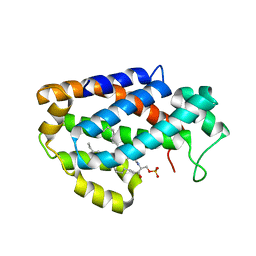 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 18:1 Ceramide-1-Phosphate (18:1-C1P) | | Descriptor: | (2S,3R,4Z)-3-hydroxy-2-[(9E)-octadec-9-enoylamino]octadec-4-en-1-yl dihydrogen phosphate, Glycolipid transfer protein domain-containing protein 1 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-04-18 | | Release date: | 2013-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
1GVS
 
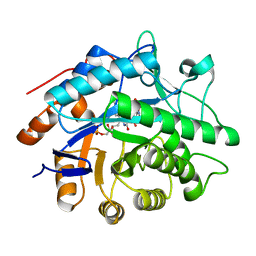 | |
1GWW
 
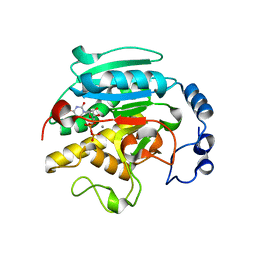 | | ALPHA-,1,3 GALACTOSYLTRANSFERASE - ALPHA-D-GLUCOSE COMPLEX | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Boix, E, Zhang, Y, Swaminathan, G.J, Brew, K, Acharya, K.R. | | Deposit date: | 2002-03-26 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Ordered Binding of Donor and Acceptor Substrates to the Retaining Glycosyltransferase, Alpha -1,3 Galactosyltransferase
J.Biol.Chem., 277, 2002
|
|
1GXY
 
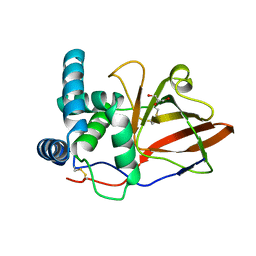 | |
8BTZ
 
 | |
1H01
 
 | | CDK2 in complex with a disubstituted 2, 4-bis anilino pyrimidine CDK4 inhibitor | | Descriptor: | (2R)-1-[4-({4-[(2,5-DICHLOROPHENYL)AMINO]PYRIMIDIN-2-YL}AMINO)PHENOXY]-3-(DIMETHYLAMINO)PROPAN-2-OL, (2S)-1-[4-({4-[(2,5-DICHLOROPHENYL)AMINO]PYRIMIDIN-2-YL}AMINO)PHENOXY]-3-(DIMETHYLAMINO)PROPAN-2-OL, CYCLIN-DEPENDENT KINASE 2, ... | | Authors: | Breault, G.A, Ellston, R.P.A, Green, S, James, S.R, Jewsbury, P.J, Midgley, C.J, Minshull, C.A, Pauptit, R.A, Tucker, J.A, Pease, J.E. | | Deposit date: | 2002-06-10 | | Release date: | 2003-07-11 | | Last modified: | 2011-10-19 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Cyclin-Dependent Kinase 4 Inhibitors as a Treatment for Cancer. Part 1: Identification and Optimisation of Substituted 4,6-Bis Anilino Pyrimidines
Bioorg.Med.Chem.Lett., 13, 2003
|
|
2NZJ
 
 | | The crystal structure of REM1 in complex with GDP | | Descriptor: | CHLORIDE ION, GTP-binding protein REM 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Turnbull, A.P, Papagrigoriou, E, Ugochukwu, E, Elkins, J.M, Soundararajan, M, Yang, X, Gorrec, F, Umeano, C, Salah, E, Burgess, N, Johansson, C, Berridge, G, Gileadi, O, Bray, J, Marsden, B, Watts, S, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C.H, Sundstrom, M, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-23 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of REM1 in complex with GDP
To be Published
|
|
1H51
 
 | | Oxidised Pentaerythritol Tetranitrate Reductase (SCN complex) | | Descriptor: | FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE, THIOCYANATE ION | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2001-05-17 | | Release date: | 2003-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Pentaerythritol Tetranitrate Reductase: "Flipped" Binding Geometries for Steroid Substrates in Different Redox States of the Enzyme
J.Mol.Biol., 310, 2001
|
|
6BWH
 
 | |
