4HQP
 
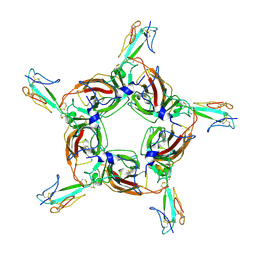 | | Alpha7 nicotinic receptor chimera and its complex with Alpha bungarotoxin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-bungarotoxin isoform V31, ... | | Authors: | Li, S.X, Cheng, K, Gomoto, R, Bren, N, Huang, S, Sine, S, Chen, L. | | Deposit date: | 2012-10-25 | | Release date: | 2013-07-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structural principles for Alpha-neurotoxin binding to and selectivity among nicotinic receptors
To be Published
|
|
4HZR
 
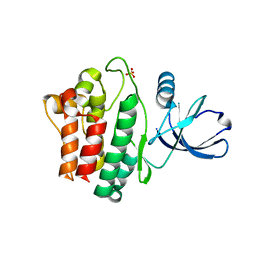 | | Crystal structure of Ack1 kinase domain | | Descriptor: | 1,2-ETHANEDIOL, Activated CDC42 kinase 1, CHLORIDE ION, ... | | Authors: | Gajiwala, K.S. | | Deposit date: | 2012-11-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Ack1: activation and regulation by allostery.
Plos One, 8, 2013
|
|
3T1E
 
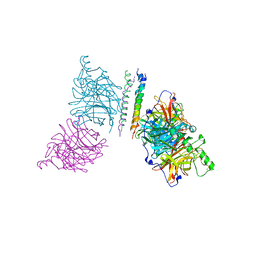 | | The structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a 4-helix bundle stalk | | Descriptor: | Hemagglutinin-neuraminidase | | Authors: | Yuan, P, Swanson, K, Leser, G.P, Paterson, R.G, Lamb, R.A, Jardetzky, T.S. | | Deposit date: | 2011-07-21 | | Release date: | 2011-09-07 | | Last modified: | 2011-09-21 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4I59
 
 | |
7ZN9
 
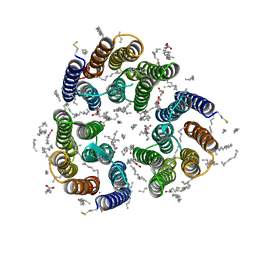 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the M state at pH 7.0 in the presence of sodium at 100K | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZNB
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the M state at pH 5.2 in the presence of sodium at 100K | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4I58
 
 | |
2UUC
 
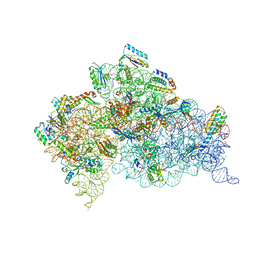 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a Valine-ASL with cmo5U in position 34 bound to an mRNA with a GUA-codon in the A-site and paromomycin. | | Descriptor: | 16S Ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Weixlbaumer, A, Murphy, F.V, Dziergowska, A, Malkiewicz, A, Vendeix, F.A.P, Agris, P.F, Ramakrishnan, V. | | Deposit date: | 2007-03-01 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines.
Nat. Struct. Mol. Biol., 14, 2007
|
|
2UU9
 
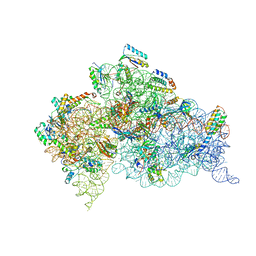 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a Valine-ASL with cmo5U in position 34 bound to an mRNA with a GUG-codon in the A-site and paromomycin. | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Weixlbaumer, A, Murphy, F.V, Dziergowska, A, Malkiewicz, A, Vendeix, F.A.P, Agris, P.F, Ramakrishnan, V. | | Deposit date: | 2007-03-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines.
Nat. Struct. Mol. Biol., 14, 2007
|
|
2UXD
 
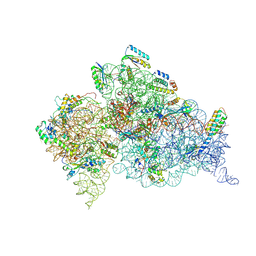 | | Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA CGGG in the context of the Thermus thermophilus 30S subunit. | | Descriptor: | 16S RIBOSOMAL RNA, A-SITE MESSENGER RNA FRAGMENT CGGG, ANTICODON STEM-LOOP OF TRANSFER RNA WITH ANTICODON CCCG, ... | | Authors: | Dunham, C.M, Selmer, M, Phelps, S.S, Kelley, A.C, Suzuki, T, Joseph, S, Ramakrishnan, V. | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Trnas with an Expanded Anticodon Loop in the Decoding Center of the 30S Ribosomal Subunit.
RNA, 13, 2007
|
|
4IAP
 
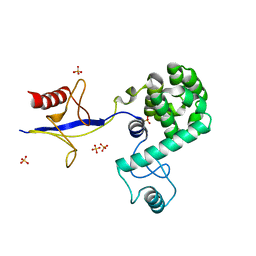 | | Crystal structure of PH domain of Osh3 from Saccharomyces cerevisiae | | Descriptor: | Oxysterol-binding protein homolog 3,Endolysin,Oxysterol-binding protein homolog 3, SULFATE ION | | Authors: | Tong, J, Im, Y.J. | | Deposit date: | 2012-12-07 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of osh3 reveals a conserved mode of phosphoinositide binding in oxysterol-binding proteins
Structure, 21, 2013
|
|
2UXB
 
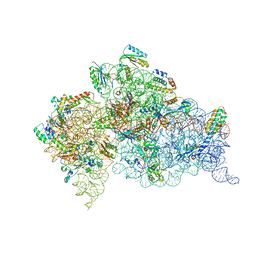 | | Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA GGGU in the context of the Thermus thermophilus 30S subunit. | | Descriptor: | 16S RIBOSOMAL RNA, A-SITE MESSENGER RNA FRAGMENT GGGU, ANTICODON STEM-LOOP OF TRANSFER RNA WITH ANTICODON ACCC, ... | | Authors: | Dunham, C.M, Selmer, M, Phelps, S.S, Kelley, A.C, Suzuki, T, Joseph, S, Ramakrishnan, V. | | Deposit date: | 2007-03-28 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of tRNAs with an expanded anticodon loop in the decoding center of the 30S ribosomal subunit.
RNA, 13, 2007
|
|
2UXC
 
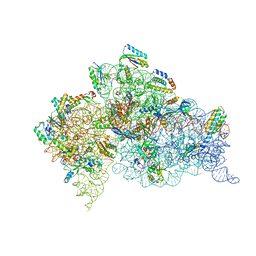 | | Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA UCGU in the context of the Thermus thermophilus 30S subunit. | | Descriptor: | 16S RIBOSOMAL RNA, A-SITE MESSENGER RNA FRAGMENT CGGG, ANTICODON STEM-LOOP OF TRANSFER RNA WITH ANTICODON ACGA, ... | | Authors: | Dunham, C.M, Selmer, M, Phelps, S.S, Kelley, A.C, Suzuki, T, Joseph, S, Ramakrishnan, V. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Trnas with an Expanded Anticodon Loop in the Decoding Center of the 30S Ribosomal Subunit.
RNA, 13, 2007
|
|
4INQ
 
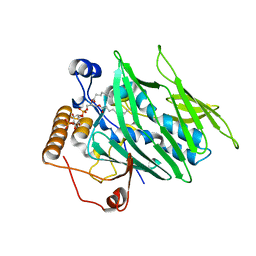 | | Crystal structure of Osh3 ORD in complex with PI(4)P from Saccharomyces cerevisiae | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, Oxysterol-binding protein homolog 3 | | Authors: | Tong, J, Im, Y.J. | | Deposit date: | 2013-01-05 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of osh3 reveals a conserved mode of phosphoinositide binding in oxysterol-binding proteins
Structure, 21, 2013
|
|
4IOV
 
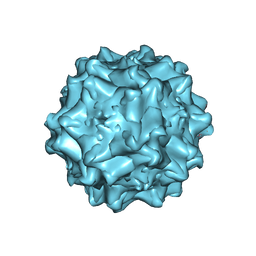 | | The structure of AAVrh32.33, a Novel Gene Delivery Vector | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Capsid protein VP1 | | Authors: | Mikalism, K, Nam, H.-J, Van vilet, K, Vandenberghe, L.H, Mays, L.E, McKenna, R, Wilson, J, Agbandje-McKenna, M. | | Deposit date: | 2013-01-08 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of AAVrh32.33, a novel gene delivery vector.
J.Struct.Biol., 186, 2014
|
|
3TF4
 
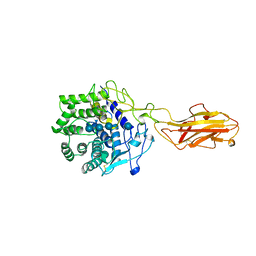 | | ENDO/EXOCELLULASE:CELLOTRIOSE FROM THERMOMONOSPORA | | Descriptor: | CALCIUM ION, T. FUSCA ENDO/EXO-CELLULASE E4 CATALYTIC DOMAIN AND CELLULOSE-BINDING DOMAIN, beta-D-glucopyranose, ... | | Authors: | Sakon, J, Wilson, D.B, Karplus, P.A. | | Deposit date: | 1997-05-30 | | Release date: | 1997-09-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca.
Nat.Struct.Biol., 4, 1997
|
|
4IC4
 
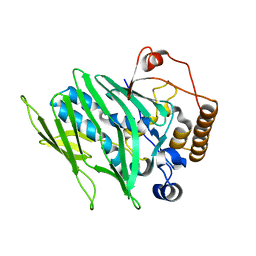 | |
4IW2
 
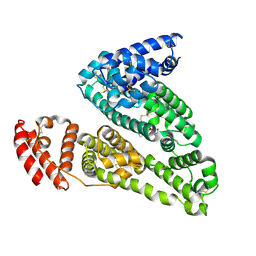 | | HSA-glucose complex | | Descriptor: | D-glucose, PHOSPHATE ION, Serum albumin, ... | | Authors: | Wang, Y, Yu, H, Shi, X, Luo, Z, Huang, M. | | Deposit date: | 2013-01-23 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural mechanism of ring-opening reaction of glucose by human serum albumin
J.Biol.Chem., 288, 2013
|
|
4IZD
 
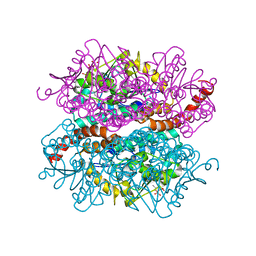 | | Crystal structure of DmdD E121A in complex with MMPA-CoA | | Descriptor: | 3-methylmercaptopropionate-CoA (MMPA-CoA), Enoyl-CoA hydratase/isomerase family protein | | Authors: | Tan, D, Tong, L. | | Deposit date: | 2013-01-29 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of DmdD, a Crotonase Superfamily Enzyme That Catalyzes the Hydration and Hydrolysis of Methylthioacryloyl-CoA.
Plos One, 8, 2013
|
|
4ISH
 
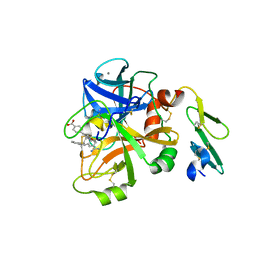 | | Structure of FACTOR VIIA in complex with the inhibitor BMS-593214 also known as 2'-[(6R,6AR,11BR)-2-CARBAMIMIDOYL-6,6A,7,11B-TETRAHYDRO-5H-INDENO[2,1-C]QUINOLIN-6-YL]-5'-HYDROXY-4'-METHOXYBIPHENYL-4-CARBOXYLIC ACID | | Descriptor: | 2'-[(6R,6aR,11bR)-2-carbamimidoyl-6,6a,7,11b-tetrahydro-5H-indeno[2,1-c]quinolin-6-yl]-5'-hydroxy-4'-methoxybiphenyl-4-carboxylic acid, CALCIUM ION, Factor VII heavy chain, ... | | Authors: | Wei, A. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery and gram-scale synthesis of BMS-593214, a potent, selective FVIIa inhibitor.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4J1X
 
 | |
4IV3
 
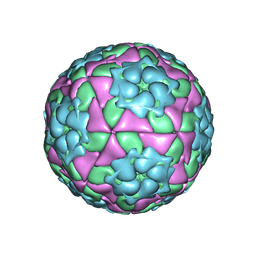 | | Crystal structure of recombinant foot-and-mouth-disease virus A22-H2093C empty capsid | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Porta, C, Kotecha, A, Burman, A, Jackson, T, Ren, J, Loureiro, S, Jones, I.M, Fry, E.E, Stuart, D.I, Charleston, B. | | Deposit date: | 2013-01-22 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Rational engineering of recombinant picornavirus capsids to produce safe, protective vaccine antigen.
Plos Pathog., 9, 2013
|
|
6VSV
 
 | |
4IOP
 
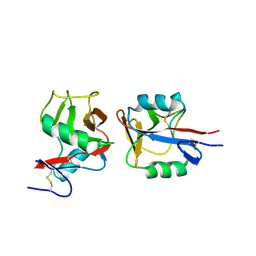 | | Crystal structure of NKp65 bound to its ligand KACL | | Descriptor: | C-type lectin domain family 2 member A, Killer cell lectin-like receptor subfamily F member 2, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, Y. | | Deposit date: | 2013-01-08 | | Release date: | 2013-07-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of NKp65 bound to its keratinocyte ligand reveals basis for genetically linked recognition in natural killer gene complex.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3FAR
 
 | |
