1ME8
 
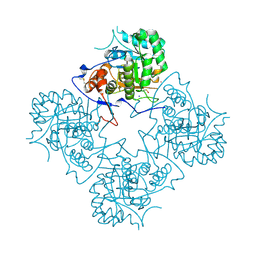 | | Inosine Monophosphate Dehydrogenase (IMPDH) From Tritrichomonas Foetus with RVP bound | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, POTASSIUM ION, RIBAVIRIN MONOPHOSPHATE, ... | | Authors: | Prosise, G.L, Wu, J, Luecke, H. | | Deposit date: | 2002-08-08 | | Release date: | 2003-01-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Tritrichomonas foetus Inosine Monophosphate Dehydrogenase
in Complex with the Inhibitor Ribavirin Monophosphate Reveals a
Catalysis-dependent Ion-binding Site
J.Biol.Chem., 277, 2002
|
|
1ME9
 
 | |
1MEA
 
 | |
1MEC
 
 | |
1MED
 
 | |
1MEE
 
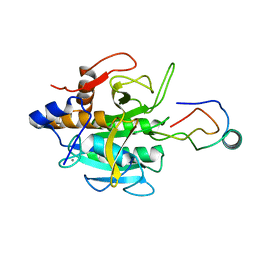 | | THE COMPLEX BETWEEN THE SUBTILISIN FROM A MESOPHILIC BACTERIUM AND THE LEECH INHIBITOR EGLIN-C | | Descriptor: | CALCIUM ION, EGLIN C, MESENTERICOPEPTIDASE | | Authors: | Dauter, Z, Betzel, C, Wilson, K.S. | | Deposit date: | 1991-04-15 | | Release date: | 1992-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex between the subtilisin from a mesophilic bacterium and the leech inhibitor eglin-C.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
1MEG
 
 | |
1MEH
 
 | |
1MEI
 
 | |
1MEJ
 
 | | Human Glycinamide Ribonucleotide Transformylase domain at pH 8.5 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Zhang, Y, Desharnais, J, Greasley, S.E, Beardsley, G.P, Boger, D.L, Wilson, I.A. | | Deposit date: | 2002-08-08 | | Release date: | 2002-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human GAR Tfase of low and high pH and with substrate beta-GAR
Biochemistry, 41, 2002
|
|
1MEK
 
 | | HUMAN PROTEIN DISULFIDE ISOMERASE, NMR, 40 STRUCTURES | | Descriptor: | PROTEIN DISULFIDE ISOMERASE | | Authors: | Kemmink, J, Darby, N.J, Dijkstra, K, Nilges, M, Creighton, T.E. | | Deposit date: | 1996-04-16 | | Release date: | 1997-04-21 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure determination of the N-terminal thioredoxin-like domain of protein disulfide isomerase using multidimensional heteronuclear 13C/15N NMR spectroscopy.
Biochemistry, 35, 1996
|
|
1MEL
 
 | | CRYSTAL STRUCTURE OF A CAMEL SINGLE-DOMAIN VH ANTIBODY FRAGMENT IN COMPLEX WITH LYSOZYME | | Descriptor: | LYSOZYME, VH SINGLE-DOMAIN ANTIBODY | | Authors: | Desmyter, A, Transue, T.R, Arbabi Gharoudi, M, Dao Thi, M, Poortmans, F, Hamers, R, Muyldermans, S, Wyns, L. | | Deposit date: | 1996-06-06 | | Release date: | 1997-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme.
Nat.Struct.Biol., 3, 1996
|
|
1MEM
 
 | |
1MEN
 
 | | complex structure of human GAR Tfase and substrate beta-GAR | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, Phosphoribosylglycinamide formyltransferase | | Authors: | Zhang, Y, Desharnais, J, Greasley, S.E, Beardsley, G.P, Boger, D.L, Wilson, I.A. | | Deposit date: | 2002-08-08 | | Release date: | 2002-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structures of human GAR Tfase of low and high pH and with substrate beta-GAR
Biochemistry, 41, 2002
|
|
1MEO
 
 | | human glycinamide ribonucleotide Transformylase at pH 4.2 | | Descriptor: | PHOSPHATE ION, Phosphoribosylglycinamide formyltransferase, SULFATE ION | | Authors: | Zhang, Y, Desharnais, J, Greasley, S.E, Beardsley, G.P, Boger, D.L, Wilson, I.A. | | Deposit date: | 2002-08-08 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures of human GAR Tfase of low and high pH and with substrate beta-GAR
Biochemistry, 41, 2002
|
|
1MEP
 
 | | Crystal Structure of Streptavidin Double Mutant S45A/D128A with Biotin: Cooperative Hydrogen-Bond Interactions in the Streptavidin-Biotin System. | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Hyre, D.E, Le Trong, I, Merritt, E.A, Stenkamp, R.E, Green, N.M, Stayton, P.S. | | Deposit date: | 2002-08-08 | | Release date: | 2003-09-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cooperative hydrogen bond interactions in the streptavidin-biotin system
Protein Sci., 15, 2006
|
|
1MEQ
 
 | | HIV gp120 C5 | | Descriptor: | Exterior Membrane Glycoprotein (GP120) | | Authors: | Caffrey, M, Jacobs, A, Guilhaudis, L. | | Deposit date: | 2002-08-08 | | Release date: | 2002-12-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the HIV gp120 C5 Domain
Eur.J.Biochem., 269, 2002
|
|
1MER
 
 | | HIV-1 MUTANT (I84V) PROTEASE COMPLEXED WITH DMP450 | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-1,3-BIS([(3-AMINO)PHENYL]METHYL)-4,7-BIS(PHENYLMETHYL)-2H-1,3-DIAZEPINONE | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with cyclic urea inhibitors.
Biochemistry, 36, 1997
|
|
1MES
 
 | | HIV-1 MUTANT (I84V) PROTEASE COMPLEXED WITH DMP323 | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with cyclic urea inhibitors.
Biochemistry, 36, 1997
|
|
1MET
 
 | | HIV-1 MUTANT (V82F) PROTEASE COMPLEXED WITH DMP323 | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with cyclic urea inhibitors.
Biochemistry, 36, 1997
|
|
1MEU
 
 | | HIV-1 MUTANT (V82F, I84V) PROTEASE COMPLEXED WITH DMP323 | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with cyclic urea inhibitors.
Biochemistry, 36, 1997
|
|
1MEW
 
 | |
1MEX
 
 | |
1MEY
 
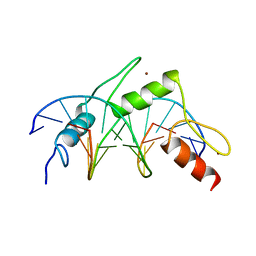 | | CRYSTAL STRUCTURE OF A DESIGNED ZINC FINGER PROTEIN BOUND TO DNA | | Descriptor: | CHLORIDE ION, CONSENSUS ZINC FINGER, DNA (5'-D(*AP*TP*GP*AP*GP*GP*CP*AP*GP*AP*AP*CP*T)-3'), ... | | Authors: | Kim, C.A, Berg, J.M. | | Deposit date: | 1996-09-27 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A 2.2 A Resolution Crystal Structure of a Designed Zinc Finger Protein Bound to DNA
Nat.Struct.Biol., 3, 1996
|
|
1MEZ
 
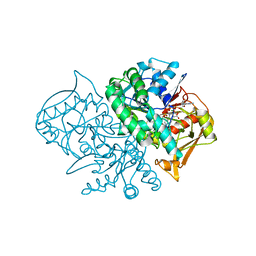 | | Structure of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase Complexed with SAMP, GDP, SO4(2-), and Mg(2+) | | Descriptor: | 2-[9-(3,4-DIHYDROXY-5-PHOSPHONOOXYMETHYL-TETRAHYDRO-FURAN-2-YL)-9H-PURIN-6-YLAMINO]-SUCCINIC ACID, Adenylosuccinate Synthetase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Iancu, C.V, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2002-08-09 | | Release date: | 2002-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Feedback inhibition and product complexes of recombinant mouse muscle adenylosuccinate synthetase.
J.Biol.Chem., 277, 2002
|
|
