1HT4
 
 | |
1QHK
 
 | |
1QQV
 
 | |
1C9Q
 
 | |
1BV8
 
 | |
1ROQ
 
 | |
1GM0
 
 | | A Form of the Pheromone-Binding Protein from Bombyx mori | | Descriptor: | PHEROMONE-BINDING PROTEIN | | Authors: | Horst, R, Damberger, F, Guntert, P, Luginbuhl, P, Nikonova, L, Peng, G, Leal, W.S, Wuthrich, K. | | Deposit date: | 2001-09-05 | | Release date: | 2001-11-30 | | Last modified: | 2018-01-17 | | Method: | SOLUTION NMR | | Cite: | NMR Structure Reveals Novel Intramolecular Regulation Mechanism for Pheromone-Binding and Release
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1HA8
 
 | |
1J3G
 
 | | Solution structure of Citrobacter Freundii AmpD | | Descriptor: | AmpD protein, ZINC ION | | Authors: | Liepinsh, E, Genereux, C, Dehareng, D, Joris, B, Otting, G. | | Deposit date: | 2003-01-31 | | Release date: | 2003-02-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Citrobacter freundii AmpD, Comparison with Bacteriophage T7 Lysozyme
and Homology with PGRP Domains
J.Mol.Biol., 327, 2003
|
|
1IMI
 
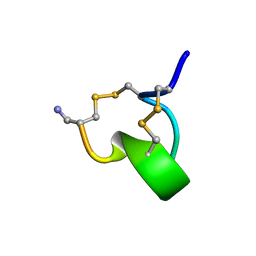 | | SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 | | Descriptor: | PROTEIN (ALPHA-CONOTOXIN IMI) | | Authors: | Maslennikov, I.V, Shenkarev, Z.O, Zhmak, M.N, Tsetlin, V.I, Ivanov, V.T, Arseniev, A.S. | | Deposit date: | 1998-11-27 | | Release date: | 1999-04-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR spatial structure of alpha-conotoxin ImI reveals a common scaffold in snail and snake toxins recognizing neuronal nicotinic acetylcholine receptors.
FEBS Lett., 444, 1999
|
|
1BFM
 
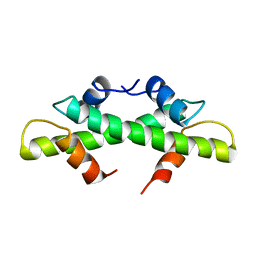 | | HISTONE B FROM METHANOTHERMUS FERVIDUS | | Descriptor: | HISTONE B | | Authors: | Starich, M.R, Sandman, K, Reeve, J.N, Summers, M.F. | | Deposit date: | 1995-09-28 | | Release date: | 1996-01-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of HMfB from the hyperthermophile, Methanothermus fervidus, confirms that this archaeal protein is a histone.
J.Mol.Biol., 255, 1996
|
|
1CR8
 
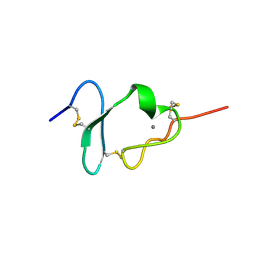 | |
1DLZ
 
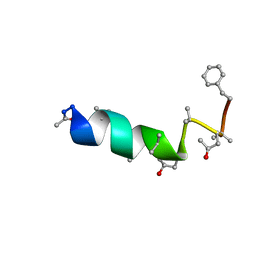 | | SOLUTION STRUCTURE OF THE CHANNEL-FORMER ZERVAMICIN IIB (PEPTAIBOL ANTIBIOTIC) | | Descriptor: | ZERVAMICIN IIB | | Authors: | Balashova, T.A, Shenkarev, Z.O, Tagaev, A.A, Ovchinnikova, T.V, Raap, J, Arseniev, A.S. | | Deposit date: | 1999-12-13 | | Release date: | 2000-02-03 | | Last modified: | 2012-12-12 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Channel-Former Zervamicin Iib in Isotropic Solvents.
FEBS Lett., 466, 2000
|
|
1D8Z
 
 | | SOLUTION STRUCTURE OF THE FIRST RNA-BINDING DOMAIN (RBD1) OF HU ANTIGEN C (HUC) | | Descriptor: | HU ANTIGEN C | | Authors: | Inoue, M, Muto, Y, Sakamoto, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-26 | | Release date: | 2000-04-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR studies on functional structures of the AU-rich element-binding domains of Hu antigen C.
Nucleic Acids Res., 28, 2000
|
|
1D9A
 
 | | SOLUTION STRUCTURE OF THE SECOND RNA-BINDING DOMAIN (RBD2) OF HU ANTIGEN C (HUC) | | Descriptor: | HU ANTIGEN C | | Authors: | Inoue, M, Muto, Y, Sakamoto, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-26 | | Release date: | 2000-04-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR studies on functional structures of the AU-rich element-binding domains of Hu antigen C.
Nucleic Acids Res., 28, 2000
|
|
2ALC
 
 | | ETHANOL REGULON TRANSCRIPTIONAL ACTIVATOR DNA-BINDING DOMAIN FROM ASPERGILLUS NIDULANS | | Descriptor: | PROTEIN (ETHANOL REGULON TRANSCRIPTIONAL ACTIVATOR), ZINC ION | | Authors: | Cerdan, R, Cahuzac, B, Felenbok, B, Guittet, E. | | Deposit date: | 1999-01-20 | | Release date: | 2000-01-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of AlcR (1-60) provides insight in the unusual DNA binding properties of this zinc binuclear cluster protein.
J.Mol.Biol., 295, 2000
|
|
2B4N
 
 | |
2AGM
 
 | | Solution structure of the R-module from AlgE4 | | Descriptor: | Poly(beta-D-mannuronate) C5 epimerase 4 | | Authors: | Aachmann, F.L, Svanem, B.I, Guntert, P, Petersen, S.B, Valla, S, Wimmer, R. | | Deposit date: | 2005-07-27 | | Release date: | 2006-01-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the R-module: a parallel beta-roll subunit from an Azotobacter vinelandii mannuronan C-5 epimerase.
J.Biol.Chem., 281, 2006
|
|
2BAI
 
 | | The Zinc finger domain of Mengovirus Leader polypeptide | | Descriptor: | Genome polyprotein, ZINC ION | | Authors: | Cornilescu, C.C, Porter, F.W, Qin, Z, Lee, M.S, Palmenberg, A.C, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-14 | | Release date: | 2006-01-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the mengovirus Leader protein zinc-finger domain.
Febs Lett., 582, 2008
|
|
1ZFI
 
 | | Solution structure of the leech carboxypeptidase inhibitor | | Descriptor: | Metallocarboxypeptidase inhibitor | | Authors: | Arolas, J.L, D'Silva, L, Popowicz, G.M, Aviles, F.X, Holak, T.A, Ventura, S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-09-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structural characterization and computational predictions of the major intermediate in oxidative folding of leech carboxypeptidase inhibitor
STRUCTURE, 13, 2005
|
|
1ZFL
 
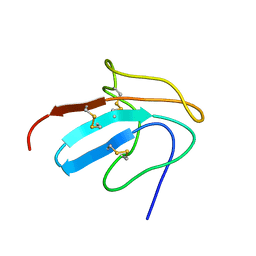 | | Solution structure of III-A, the major intermediate in the oxidative folding of leech carboxypeptidase inhibitor | | Descriptor: | Metallocarboxypeptidase inhibitor | | Authors: | Arolas, J.L, D'Silva, L, Popowicz, G.M, Aviles, F.X, Holak, T.A, Ventura, S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-09-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structural characterization and computational predictions of the major intermediate in oxidative folding of leech carboxypeptidase inhibitor
STRUCTURE, 13, 2005
|
|
2F40
 
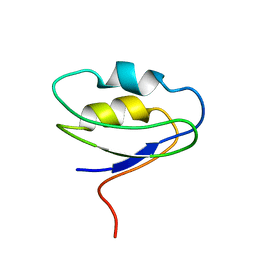 | |
1TU2
 
 | | THE COMPLEX OF NOSTOC CYTOCHROME F AND PLASTOCYANIN DETERMIN WITH PARAMAGNETIC NMR. BASED ON THE STRUCTURES OF CYTOCHROME F AND PLASTOCYANIN, 10 STRUCTURES | | Descriptor: | Apocytochrome f, COPPER (II) ION, HEME C, ... | | Authors: | Diaz-Moreno, I, Diaz-Quintana, A, De la Rosa, M.A, Ubbink, M. | | Deposit date: | 2004-06-24 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the complex between plastocyanin and cytochrome f from the cyanobacterium Nostoc sp. PCC 7119 as determined by paramagnetic NMR. The balance between electrostatic and hydrophobic interactions within the transient complex determines the relative orientation of the two proteins.
J.Biol.Chem., 280, 2005
|
|
2ETZ
 
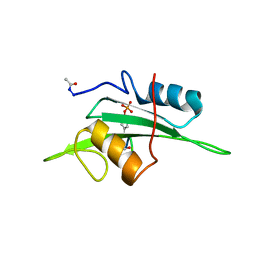 | | The NMR minimized average structure of the Itk SH2 domain bound to a phosphopeptide | | Descriptor: | Lymphocyte cytosolic protein 2 phosphopeptide fragment, Tyrosine-protein kinase ITK/TSK | | Authors: | Sundd, M, Pletneva, E.V, Fulton, D.B, Andreotti, A.H. | | Deposit date: | 2005-10-27 | | Release date: | 2006-02-07 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Molecular Details of Itk Activation by Prolyl Isomerization and Phospholigand Binding: The NMR Structure of the Itk SH2 Domain Bound to a Phosphopeptide.
J.Mol.Biol., 357, 2006
|
|
2EU0
 
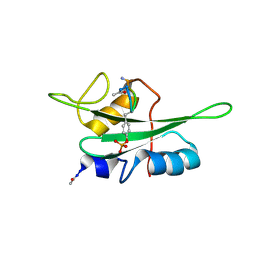 | | The NMR ensemble structure of the Itk SH2 domain bound to a phosphopeptide | | Descriptor: | Lymphocyte cytosolic protein 2 phosphopeptide fragment, Tyrosine-protein kinase ITK/TSK | | Authors: | Sundd, M, Pletneva, E.V, Fulton, D.B, Andreotti, A.H. | | Deposit date: | 2005-10-27 | | Release date: | 2006-02-07 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Molecular Details of Itk Activation by Prolyl Isomerization and Phospholigand Binding: The NMR Structure of the Itk SH2 Domain Bound to a Phosphopeptide.
J.Mol.Biol., 357, 2006
|
|
