1NZ0
 
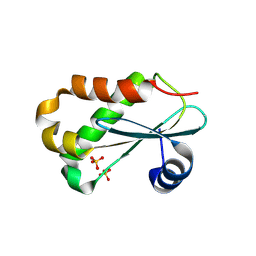 | | RNASE P PROTEIN FROM THERMOTOGA MARITIMA | | Descriptor: | Ribonuclease P protein component, SULFATE ION | | Authors: | Kazantsev, A.V, Krivenko, A.A, Harrington, D.J, Carter, R.J, Holbrook, S.R, Adams, P.D, Pace, N.R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-02-14 | | Release date: | 2003-06-24 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution structure of RNase P protein from Thermotoga maritima.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
6KHS
 
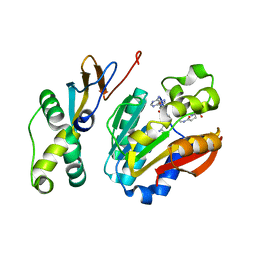 | |
6KM2
 
 | | Human Carbonic Anhydrase II V143I variant 15 atm CO2 | | Descriptor: | BICARBONATE ION, CARBON DIOXIDE, Carbonic anhydrase 2, ... | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2019-07-30 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structural insights into the effect of active-site mutation on the catalytic mechanism of carbonic anhydrase.
Iucrj, 7, 2020
|
|
6KNK
 
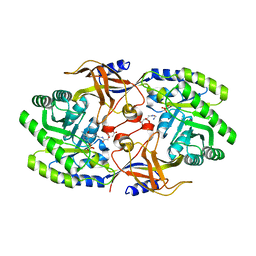 | | Crystal structure of SbnH in complex with citryl-diaminoethane | | Descriptor: | (2S)-2-{2-[(2-AMINOETHYL)AMINO]-2-OXOETHYL}-2-HYDROXYBUTANEDIOIC ACID, (2~{S})-2-[2-[2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]ethylamino]-2-oxidanylidene-ethyl]-2-oxidanyl-butanedioic acid, PHOSPHATE ION, ... | | Authors: | Tang, J, Ju, Y, Zhou, H. | | Deposit date: | 2019-08-05 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into Substrate Recognition and Activity Regulation of the Key Decarboxylase SbnH in Staphyloferrin B Biosynthesis.
J.Mol.Biol., 431, 2019
|
|
5LZN
 
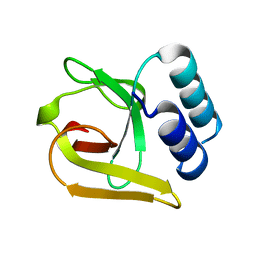 | | -TIP microtubule-binding domain | | Descriptor: | Calmodulin-regulated spectrin-associated protein 3 | | Authors: | Stangier, M.M, Steinmetz, M.O. | | Deposit date: | 2016-09-30 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A structural model for microtubule minus-end recognition and protection by CAMSAP proteins.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6RRP
 
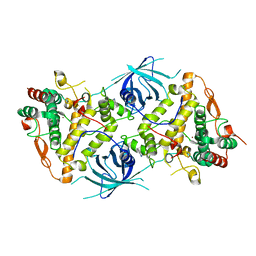 | | Crystal structure of tyrosinase PvdP from Pseudomonas aeruginosa bound to copper and phenylthiourea | | Descriptor: | COPPER (II) ION, N-PHENYLTHIOUREA, PvdP | | Authors: | Wibowo, J.P, Batista, F.A, van Oosterwijk, N, Groves, M.R, Dekker, F.J, Quax, W.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel mechanism of inhibition by phenylthiourea on PvdP, a tyrosinase synthesizing pyoverdine of Pseudomonas aeruginosa.
Int.J.Biol.Macromol., 146, 2020
|
|
2V1Y
 
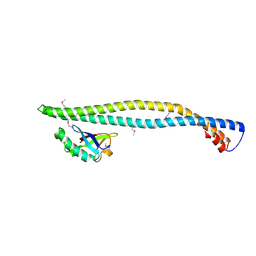 | | Structure of a phosphoinositide 3-kinase alpha adaptor-binding domain (ABD) in a complex with the iSH2 domain from p85 alpha | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY SUBUNIT ALPHA, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT ALPHA ISOFORM | | Authors: | Miled, N, Yan, Y, Hon, W.C, Perisic, O, Zvelebil, M, Inbar, Y, Schneidman-Duhovny, D, Wolfson, H.J, Backer, J.M, Williams, R.L. | | Deposit date: | 2007-05-30 | | Release date: | 2007-07-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of Two Classes of Cancer Mutations in the Phosphoinositide 3-Kinase Catalytic Subunit.
Science, 317, 2007
|
|
6KIV
 
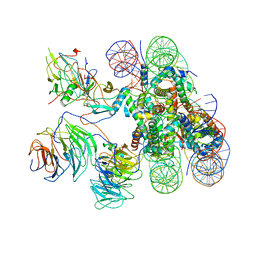 | | Cryo-EM structure of human MLL1-ubNCP complex (4.0 angstrom) | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
4WPY
 
 | | Racemic crystal structure of Rv1738 from Mycobacterium tuberculosis (Form-II) | | Descriptor: | GLYCEROL, protein DL-Rv1738, trifluoroacetic acid | | Authors: | Bunker, R.D, Mandal, K, Kent, S.B.H, Baker, E.N. | | Deposit date: | 2014-10-21 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A functional role of Rv1738 in Mycobacterium tuberculosis persistence suggested by racemic protein crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2VA9
 
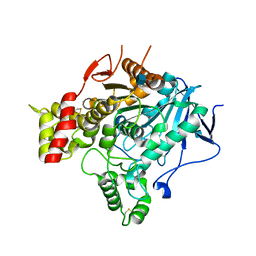 | | Structure of native TcAChE after a 9 seconds annealing to room temperature during the first 5 seconds of which laser irradiation at 266nm took place | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE | | Authors: | Colletier, J.-P, Sanson, B, Royant, A, Specht, A, Nachon, F, Masson, P, Zaccai, G, Sussman, J.L, Goeldner, M, Silman, I, Bourgeois, D, Weik, M. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Use of a 'Caged' Analog to Study Traffic of Choline within Acetylcholinesterase by Kinetic Crystallography
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2V7G
 
 | |
2VDK
 
 | | Re-refinement of Integrin AlphaIIbBeta3 Headpiece | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Springer, T.A, Zhu, J, Xiao, T. | | Deposit date: | 2007-10-10 | | Release date: | 2008-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Distinctive Recognition of Fibrinogen Gammac Peptide by the Platelet Integrin Alphaiibbeta3.
J.Cell Biol., 182, 2008
|
|
1O71
 
 | | Crystal structure of the water-soluble state of the pore-forming cytolysin Sticholysin II complexed with glycerol | | Descriptor: | GLYCEROL, STICHOLYSIN II | | Authors: | Mancheno, J.M, Martinez-Ripoll, M, Gavilanes, J.G, Hermoso, J.A. | | Deposit date: | 2002-10-23 | | Release date: | 2003-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal and Electron Microscopy Structures of Sticholysin II Actinoporin Reveal Insights Into the Mechanism of Membrane Pore Formation
Structure, 11, 2003
|
|
2V2B
 
 | |
4WLR
 
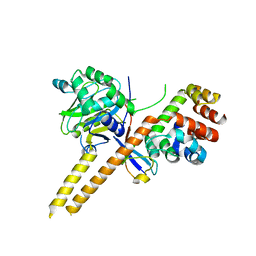 | | Crystal Structure of mUCH37-hRPN13 CTD-hUb complex | | Descriptor: | Polyubiquitin-B, Proteasomal ubiquitin receptor ADRM1, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Hemmis, C.W, Hill, C.P, VanderLinden, R, Whitby, F.G. | | Deposit date: | 2014-10-07 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural Basis for the Activation and Inhibition of the UCH37 Deubiquitylase.
Mol.Cell, 57, 2015
|
|
2VDO
 
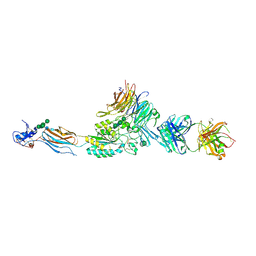 | | Integrin AlphaIIbBeta3 Headpiece Bound to Fibrinogen Gamma chain peptide, HHLGGAKQAGDV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FIBRINOGEN, ... | | Authors: | Springer, T.A, Zhu, J, Xiao, T. | | Deposit date: | 2007-10-10 | | Release date: | 2008-09-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural Basis for Distinctive Recognition of Fibrinogen Gammac Peptide by the Platelet Integrin Alphaiibbeta3.
J.Cell Biol., 182, 2008
|
|
2VE7
 
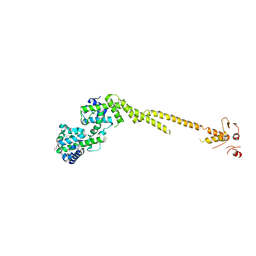 | | Crystal structure of a bonsai version of the human Ndc80 complex | | Descriptor: | GLYCEROL, KINETOCHORE PROTEIN HEC1, KINETOCHORE PROTEIN SPC25, ... | | Authors: | Ciferri, C, Pasqualato, S, Dos Reis, G, Screpanti, E, Maiolica, A, Polka, J, De Luca, J.G, De Wulf, P, Salek, M, Rappsilber, J, Moores, C.A, Salmon, E.D, Musacchio, A. | | Deposit date: | 2007-10-17 | | Release date: | 2008-05-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Implications for Kinetochore-Microtubule Attachment from the Structure of an Engineered Ndc80 Complex
Cell(Cambridge,Mass.), 133, 2008
|
|
2VFT
 
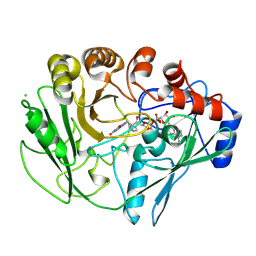 | |
2NOL
 
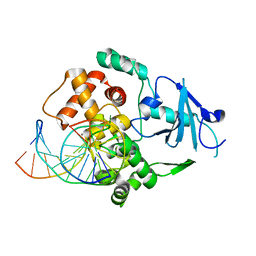 | | Structure of catalytically inactive human 8-oxoguanine glycosylase distal crosslink to oxoG DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Banerjee, A, Radom, C.T, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
6JTG
 
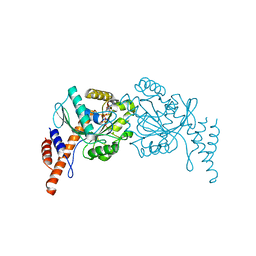 | |
6JU1
 
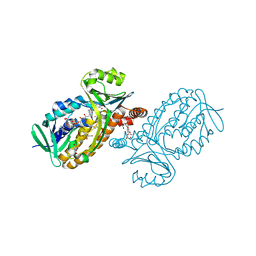 | | p-Hydroxybenzoate hydroxylase Y385F mutant complexed with 3,4-dihydroxybenzoate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 4-hydroxybenzoate 3-monooxygenase, ... | | Authors: | Yato, M, Arakawa, T, Yamada, C, Fushinobu, S. | | Deposit date: | 2019-04-12 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Understanding the Molecular Mechanism Underlying the High Catalytic Activity ofp-Hydroxybenzoate Hydroxylase Mutants for Producing Gallic Acid.
Biochemistry, 58, 2019
|
|
4WUB
 
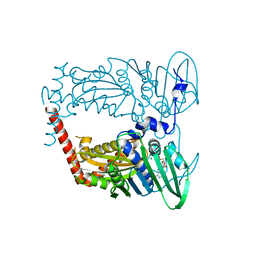 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from 100 mM KCl condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2014-10-31 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6K17
 
 | | Crystal structure of EXD2 exonuclease domain | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, SODIUM ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
6K1E
 
 | | Crystal structure of EXD2 exonuclease domain soaked in Mg and GMP | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, MAGNESIUM ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
4WUZ
 
 | | Crystal structure of lambda exonuclease in complex with DNA and Ca2+ | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*T*TP*CP*GP*GP*TP*AP*CP*AP*GP*TP*AP*G)-3'), DNA (5'-D(P*AP*GP*CP*TP*AP*CP*TP*GP*TP*AP*CP*CP*GP*A)-3'), ... | | Authors: | Zhang, J, Bell, C.E. | | Deposit date: | 2014-11-04 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of lambda Exonuclease in Complex with DNA and Ca(2+).
Biochemistry, 53, 2014
|
|
