1PVF
 
 | | E.coli IPP isomerase in complex with diphosphate | | Descriptor: | DIPHOSPHATE, Isopentenyl-diphosphate delta-isomerase, MAGNESIUM ION, ... | | Authors: | Wouters, J, Durisotti, V, Oudjama, Y, Stalon, V, Droogmans, L. | | Deposit date: | 2003-06-27 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | E.coli Isopentenyl Diphosphate: Dimethylallyl Diphosphate isomerase in complex with diphosphate
To be Published
|
|
4K3Q
 
 | | E. coli sliding clamp in complex with AcQLDAF | | Descriptor: | (ACE)QLDAF, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-11 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
2PQ9
 
 | | E. coli EPSPS liganded with (R)-difluoromethyl tetrahedral reaction intermediate analog | | Descriptor: | (3R,4S,5R)-5-[(1R)-1-CARBOXY-2,2-DIFLUORO-1-(PHOSPHONOOXY)ETHOXY]-4-HYDROXY-3-(PHOSPHONOOXY)CYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID | | Authors: | Healy-Fried, M.L, Funke, T, Han, H, Schonbrunn, E. | | Deposit date: | 2007-05-01 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Differential inhibition of class I and class II 5-enolpyruvylshikimate-3-phosphate synthases by tetrahedral reaction intermediate analogues.
Biochemistry, 46, 2007
|
|
3UPK
 
 | | E. cloacae MURA in complex with UNAG | | Descriptor: | 1,2-ETHANEDIOL, TRIETHYLENE GLYCOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | Authors: | Zhu, J.-Y, Yang, Y, Schonbrunn, E. | | Deposit date: | 2011-11-18 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional Consequence of Covalent Reaction of Phosphoenolpyruvate with UDP-N-acetylglucosamine 1-Carboxyvinyltransferase (MurA).
J.Biol.Chem., 287, 2012
|
|
4K3O
 
 | | E. coli sliding clamp in complex with AcQADLF | | Descriptor: | (ACE)QADLF, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-11 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
3A89
 
 | |
3A7Y
 
 | |
3A7Z
 
 | |
3A8B
 
 | |
4K3R
 
 | | E. coli sliding clamp in complex with AcQLDLA | | Descriptor: | (ACE)QLDLA, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-11 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
4K3L
 
 | | E. coli sliding clamp in complex with AcLF dipeptide | | Descriptor: | 1,2-ETHANEDIOL, ACETYL GROUP, CALCIUM ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
4K3S
 
 | | E. coli sliding clamp in P1 crystal space group | | Descriptor: | CALCIUM ION, DNA polymerase III subunit beta, O-ACETALDEHYDYL-HEXAETHYLENE GLYCOL, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-11 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
2XSK
 
 | | E. coli curli protein CsgC - SeCys | | Descriptor: | ACETATE ION, CSGC | | Authors: | Salgado, P.S, Taylor, J.D, Cota, E, Matthews, S.J. | | Deposit date: | 2010-09-29 | | Release date: | 2010-12-29 | | Last modified: | 2014-01-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extending the Usability of the Phasing Power of Diselenide Bonds: Secys Sad Phasing of Csgc Using a Non-Auxotrophic Strain.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3A84
 
 | |
6S1S
 
 | | Crystal structure of AmpC from Pseudomonas aeruginosa in complex with [3-(2-carboxyvinyl)phenyl]boronic acid] inhibitor | | Descriptor: | (~{E})-3-[3-(dihydroxyboranyl)phenyl]prop-2-enoic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Kekez, I, Vicario, M, Bellio, P, Tosoni, E, Celenza, G, Blazquez, J, Tondi, D, Cendron, L. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Phenylboronic Acids Probing Molecular Recognition against Class A and Class C beta-lactamases.
Antibiotics, 8, 2019
|
|
2B76
 
 | | E. coli Quinol fumarate reductase FrdA E49Q mutation | | Descriptor: | CITRATE ANION, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Maklashina, E, Iverson, T.M, Sher, Y, Kotlyar, V, Mirza, O, Andrell, J, Hudson, J.M, Armstrong, F.A, Cecchini, G. | | Deposit date: | 2005-10-03 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Fumarate Reductase and Succinate Oxidase Activity of Escherichia coli Complex II Homologs Are Perturbed Differently by Mutation of the Flavin Binding Domain
J.Biol.Chem., 281, 2006
|
|
4K3K
 
 | |
1SKU
 
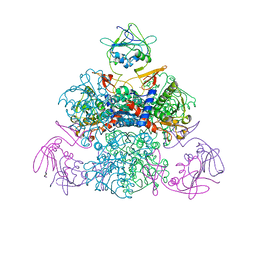 | | E. coli Aspartate Transcarbamylase 240's Loop Mutant (K244N) | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, MALONATE ION, ... | | Authors: | Alam, N, Stieglitz, K.A, Caban, M.D, Gourinath, S, Tsuruta, H, Kantrowitz, E.R. | | Deposit date: | 2004-03-05 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 240s Loop Interactions Stabilize the T State of Escherichia coli Aspartate Transcarbamoylase.
J.Biol.Chem., 279, 2004
|
|
4FYW
 
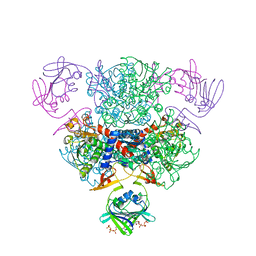 | | E. coli Aspartate Transcarbamoylase complexed with CTP | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Cockrell, G.M, Kantrowitz, E.R. | | Deposit date: | 2012-07-05 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Metal Ion Involvement in the Allosteric Mechanism of Escherichia coli Aspartate Transcarbamoylase.
Biochemistry, 51, 2012
|
|
4DUV
 
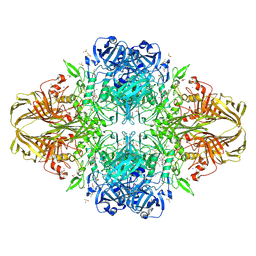 | | E. coli (lacZ) beta-galactosidase (G974A) 2-deoxy-galactosyl-enzyme and bis-Tris complex | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-deoxy-alpha-D-galactopyranose, Beta-galactosidase, ... | | Authors: | Wheatley, R.W, Lo, S, Janzcewicz, L.J, Dugdale, M.L, Huber, R.E. | | Deposit date: | 2012-02-22 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Glucose Acceptor site of lacZ beta-galactosidase for the synthesis of allolactose - the natural inducer of the lac operon
To be Published
|
|
4FYY
 
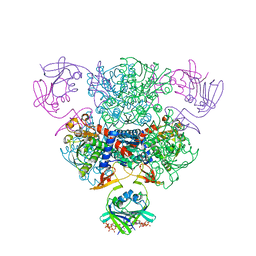 | | E. coli Aspartate Transcarbamoylase Complexed with CTP, UTP, and Mg2+ | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Cockrell, G.M, Kantrowitz, E.R. | | Deposit date: | 2012-07-05 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9395 Å) | | Cite: | Metal Ion Involvement in the Allosteric Mechanism of Escherichia coli Aspartate Transcarbamoylase.
Biochemistry, 51, 2012
|
|
3SWD
 
 | | E. coli MurA in complex with UDP-N-acetylmuramic acid and covalent adduct of PEP with Cys115 | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-07-13 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional Consequence of Covalent Reaction of Phosphoenolpyruvate with UDP-N-acetylglucosamine 1-Carboxyvinyltransferase (MurA).
J.Biol.Chem., 287, 2012
|
|
4FYX
 
 | | E. coli Aspartate Transcarbamoylase complexed with dCTP, UTP, and Mg2+ | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, ... | | Authors: | Cockrell, G.M, Kantrowitz, E.R. | | Deposit date: | 2012-07-05 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0889 Å) | | Cite: | Metal Ion Involvement in the Allosteric Mechanism of Escherichia coli Aspartate Transcarbamoylase.
Biochemistry, 51, 2012
|
|
3CIR
 
 | | E. coli Quinol fumarate reductase FrdA T234A mutation | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tomasiak, T.M, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2008-03-11 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | A threonine on the active site loop controls transition state formation in Escherichia coli respiratory complex II.
J.Biol.Chem., 283, 2008
|
|
3F1V
 
 | | E. coli Beta Sliding Clamp, 148-153 Ala Mutant | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA polymerase III subunit beta | | Authors: | Cody, V. | | Deposit date: | 2008-10-28 | | Release date: | 2009-09-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Sliding clamp-DNA interactions are required for viability and contribute to DNA polymerase management in Escherichia coli.
J.Mol.Biol., 387, 2009
|
|
