8XF2
 
 | | Crystal structure of the reassembled C-phycocyanin hexamer from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHOSPHATE ION, ... | | Authors: | Nguyen, H.K, Teramoto, T, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Disassembly and reassembly of the non-conventional thermophilic C-phycocyanin.
J.Biosci.Bioeng., 137, 2024
|
|
8XF3
 
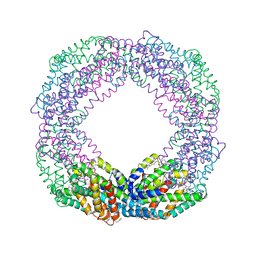 | | Crystal structure of the reassembled C-phycocyanin octamer from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHOSPHATE ION, ... | | Authors: | Nguyen, H.K, Teramoto, T, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Disassembly and reassembly of the non-conventional thermophilic C-phycocyanin.
J.Biosci.Bioeng., 137, 2024
|
|
5HTI
 
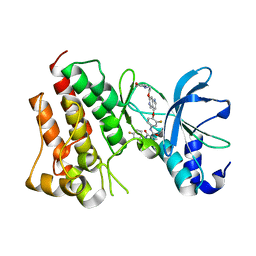 | | Crystal structure of c-Met kinase domain in complex with LXM108 | | Descriptor: | Hepatocyte growth factor receptor, N-[3-fluoro-4-({7-[2-(morpholin-4-yl)ethoxy]-1,6-naphthyridin-4-yl}oxy)phenyl]-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | Authors: | Liu, Q.F, Xu, Y.C. | | Deposit date: | 2016-01-26 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of c-Met kinase domain in complex with LXM108
to be published
|
|
7SAG
 
 | |
8S60
 
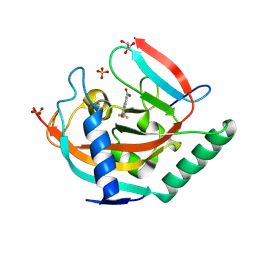 | | Tankyrase 2 in complex with a quinazolin-4-one inhibitor | | Descriptor: | 8-nitro-2-[4-(trifluoromethyl)phenyl]-3H-quinazolin-4-one, GLYCEROL, Poly [ADP-ribose] polymerase tankyrase-2, ... | | Authors: | Bosetti, C, Paakkonen, J, Lehtio, L. | | Deposit date: | 2024-02-26 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Substitutions at the C-8 position of quinazolin-4-ones improve the potency of nicotinamide site binding tankyrase inhibitors.
Eur.J.Med.Chem., 288, 2025
|
|
6YJC
 
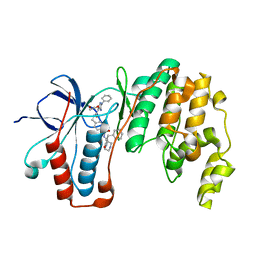 | | Crystal structure of p38alpha in complex with SR154 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-oxidanylidene-1-piperazin-1-yl-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Joerger, A.C, Schroeder, M, Roehm, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-02 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74100935 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
1E2V
 
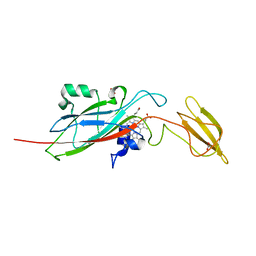 | | N153Q mutant of cytochrome f from Chlamydomonas reinhardtii | | Descriptor: | ACETATE ION, CYTOCHROME F, HEME C | | Authors: | Sainz, G, Carrell, C.J, Ponamarev, M.V, Soriano, G.M, Cramer, W.A, Smith, J.L. | | Deposit date: | 2000-05-29 | | Release date: | 2000-08-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Interruption of the Internal Water Chain of Cytochrome F Impairs Photosynthetic Function
Biochemistry, 39, 2000
|
|
1E2Z
 
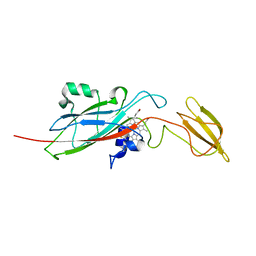 | | Q158L mutant of cytochrome f from Chlamydomonas reinhardtii | | Descriptor: | CYTOCHROME F, HEME C | | Authors: | Sainz, G, Carrell, C.J, Ponamarev, M.V, Soriano, G.M, Cramer, W.A, Smith, J.L. | | Deposit date: | 2000-05-30 | | Release date: | 2000-08-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interruption of the Internal Water Chain of Cytochrome F Impairs Photosynthetic Function
Biochemistry, 39, 2000
|
|
8W6Z
 
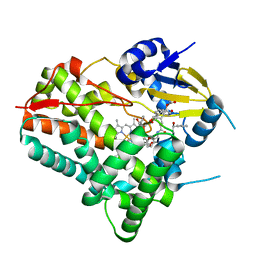 | | Substrate-bound crystal structure of a P450 enzyme DmlH that catalyze intramolecular phenol coupling in the biosynthesis of cihanmycins | | Descriptor: | (E)-3-[2-[(2R,3S)-3-[(1R)-1-aminocarbonyloxypropyl]oxiran-2-yl]phenyl]prop-2-enoic acid, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Fang, C, Zhang, L, Zhu, Y, Zhang, C. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-12 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Biosynthesis of Cihanmycins Reveal Cytochrome P450-Catalyzed Intramolecular C-O Phenol Coupling Reactions.
J.Am.Chem.Soc., 2024
|
|
4Y46
 
 | |
8HAW
 
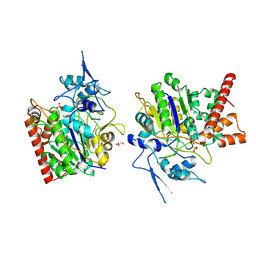 | | An auto-activation mechanism of plant non-specific phospholipase C | | Descriptor: | CALCIUM ION, GLYCEROL, Non-specific phospholipase C4, ... | | Authors: | Zhao, F, Fan, R.Y, Guan, Z.Y, Guo, L, Yin, P. | | Deposit date: | 2022-10-26 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into the mechanism of phospholipid hydrolysis by plant non-specific phospholipase C.
Nat Commun, 14, 2023
|
|
8T3J
 
 | | Crystal structure of native exfoliative toxin C (ExhC) from Mammaliicoccus sciuri | | Descriptor: | Exfoliative toxin C | | Authors: | Calil, F.A, Gismene, C, Hernandez Gonzalez, J.E, Ziem Nascimento, A.F, Santisteban, A.R.N, Arni, R.K, Barros Mariutti, R. | | Deposit date: | 2023-06-07 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Necrotic activity of ExhC from Mammaliicoccus sciuri is mediated by specific amino acid residues.
Int.J.Biol.Macromol., 254, 2024
|
|
6F4M
 
 | |
6QNO
 
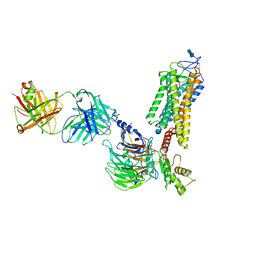 | | Rhodopsin-Gi protein complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab antibody fragment heavy chain, Fab antibody fragment light chain, ... | | Authors: | Tsai, C.-J, Marino, J, Adaixo, R.J, Pamula, F, Muehle, J, Maeda, S, Flock, T, Taylor, N.M.I, Mohammed, I, Matile, H, Dawson, R.J.P, Deupi, X, Stahlberg, H, Schertler, G.F.X. | | Deposit date: | 2019-02-11 | | Release date: | 2019-07-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.38 Å) | | Cite: | Cryo-EM structure of the rhodopsin-G alpha i-beta gamma complex reveals binding of the rhodopsin C-terminal tail to the G beta subunit.
Elife, 8, 2019
|
|
6NXK
 
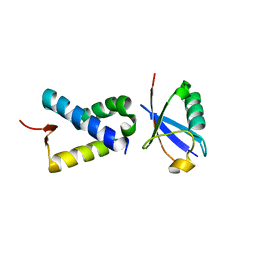 | | Ubiquitin binding variants | | Descriptor: | Anaphase-promoting complex subunit 2, Polyubiquitin-C | | Authors: | Miller, D.J, Watson, E.R. | | Deposit date: | 2019-02-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein engineering of a ubiquitin-variant inhibitor of APC/C identifies a cryptic K48 ubiquitin chain binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
9GI9
 
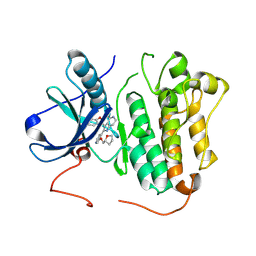 | | Wildtype EGFR bound with (R)-3-((3-chloro-2-methoxyphenyl)amino)-2-(3-((tetrahydrofuran-2-yl)methoxy)pyridin-4-yl)-1,5,6,7-tetrahydro-4H-pyrrolo[3,2-c]pyridin-4-one | | Descriptor: | 3-[(3-chloranyl-2-methoxy-phenyl)amino]-2-[3-[[(2~{R})-oxolan-2-yl]methoxy]pyridin-4-yl]-1,5,6,7-tetrahydropyrrolo[3,2-c]pyridin-4-one, Epidermal growth factor receptor | | Authors: | Pagliarini, R.A, Henderson, J.A, Borrelli, D.R, Brooijmans, N, Hilbert, B.J, Huff, M.R, Ito, T, Kryukov, G.V, Ladd, B, Martin, B.R, Milgram, B.C, Motiwala, H, OHearn, E, Wang, W, Hata, A, Bellier, J, Kuzmic, P, Guzman-Perez, A, Jackson, E.L, Stuart, D.D. | | Deposit date: | 2024-08-18 | | Release date: | 2025-05-14 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | STX-721, a Covalent EGFR/HER2 Exon 20 Inhibitor, Utilizes Exon 20-Mutant Dynamic Protein States and Achieves Unique Mutant Selectivity Across Human Cancer Models.
Clin.Cancer Res., 2025
|
|
6T6E
 
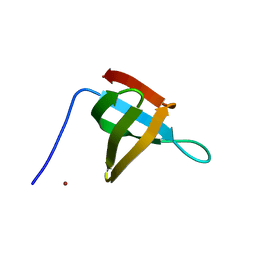 | |
8QRI
 
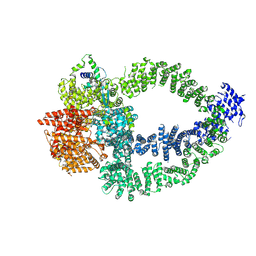 | | TRRAP and EP400 in the human Tip60 complex | | Descriptor: | E1A-binding protein p400, Transformation/transcription domain-associated protein | | Authors: | Li, C, Smirnova, E, Schnitzler, C, Crucifix, C, Concordet, J.P, Brion, A, Poterszman, A, SChultz, P, Papai, G, Ben-Shem, A. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-07 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the human TIP60-C histone exchange and acetyltransferase complex.
Nature, 635, 2024
|
|
6XX3
 
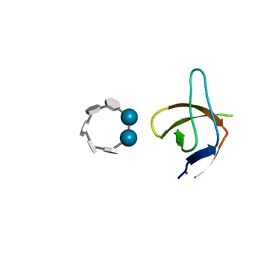 | |
4Y5H
 
 | |
6RQF
 
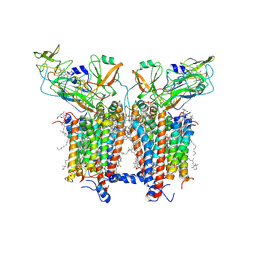 | | 3.6 Angstrom cryo-EM structure of the dimeric cytochrome b6f complex from Spinacia oleracea with natively bound thylakoid lipids and plastoquinone molecules | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Malone, L.A, Qian, P, Mayneord, G.E, Hitchcock, A, Farmer, D, Thompson, R, Swainsbury, D.J.K, Ranson, N, Hunter, C.N, Johnson, M.P. | | Deposit date: | 2019-05-15 | | Release date: | 2019-11-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Cryo-EM structure of the spinach cytochrome b6f complex at 3.6 angstrom resolution.
Nature, 575, 2019
|
|
7PPE
 
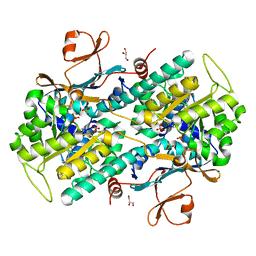 | | CRYSTAL STRUCTURE OF NAMPT IN COMPLEX WITH COMPOUND 1 | | Descriptor: | GLYCEROL, N-[4-[(4R)-4-methyl-1-(oxan-4-yl)-6-oxidanylidene-4,5-dihydropyridazin-3-yl]phenyl]-1,3-dihydropyrrolo[3,4-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Hillig, R.C. | | Deposit date: | 2021-09-13 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A Novel NAMPT Inhibitor-Based Antibody-Drug Conjugate Payload Class for Cancer Therapy.
Bioconjug.Chem., 33, 2022
|
|
9CUV
 
 | |
6Y38
 
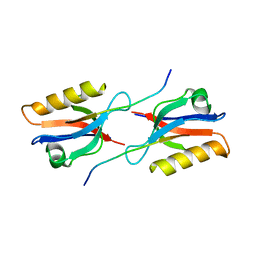 | | Crystal structure of Whirlin PDZ3 in complex with Myosin 15a C-terminal PDZ binding motif peptide | | Descriptor: | Chains: C,D, Whirlin | | Authors: | Zhu, Y, Delhommel, F, Haouz, A, Caillet-Saguy, C, Vaney, M, Mechaly, A.E, Wolff, N. | | Deposit date: | 2020-02-17 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Deciphering the Unexpected Binding Capacity of the Third PDZ Domain of Whirlin to Various Cochlear Hair Cell Partners.
J.Mol.Biol., 432, 2020
|
|
4YO7
 
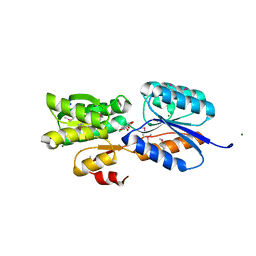 | | Crystal Structure of an ABC transporter solute binding protein (IPR025997) from Bacillus halodurans C-125 (BH2323, TARGET EFI-511484) with bound myo-inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-11 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of an ABC transporter solute binding protein (IPR025997) from Bacillus halodurans C-125 (BH2323, TARGET EFI-511484) with bound myo-inositol
To be published
|
|
