6ML5
 
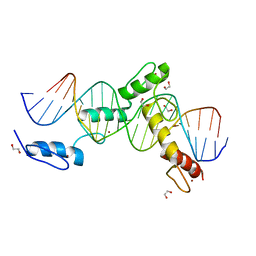 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*CP*GP*AP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*TP*CP*GP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
8SAD
 
 | |
7YBG
 
 | |
8SAE
 
 | |
7YEU
 
 | | Superfolder green fluorescent protein with phosphine unnatural amino acid P3BF | | Descriptor: | Superfolder green fluorescent protein | | Authors: | Hu, C, Duan, H.Z, Liu, X.H, Chen, Y.X, Wang, J.Y. | | Deposit date: | 2022-07-06 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Genetically Encoded Phosphine Ligand for Metalloprotein Design.
J.Am.Chem.Soc., 144, 2022
|
|
7OWH
 
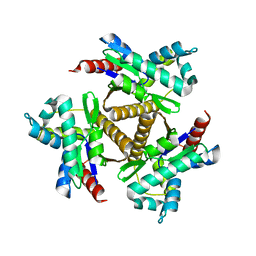 | | Odinarchaeota Adenylate kinase (OdinAK) native structure | | Descriptor: | Adenylate kinase, CHLORIDE ION | | Authors: | Aberg-Zingmark, E, Grundstrom, C, Verma, A, Wolf-Watz, M, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2021-06-18 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Insights into the evolution of enzymatic specificity and catalysis: From Asgard archaea to human adenylate kinases.
Sci Adv, 8, 2022
|
|
8UH7
 
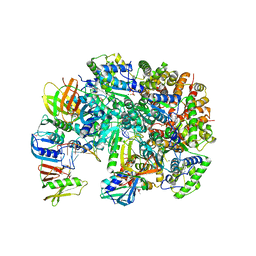 | | Structure of T4 Bacteriophage clamp loader bound to the T4 clamp, primer-template DNA, and ATP analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Primer DNA strand, ... | | Authors: | Gee, C.L, Marcus, K, Kelch, B.A, Makino, D.L. | | Deposit date: | 2023-10-07 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.628 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7OR5
 
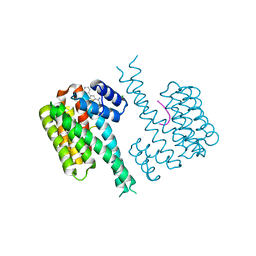 | | Ternary complex of 14-3-3 sigma, NotchpS1917 phosphopeptide, and WQ162 | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Neurogenic locus notch homolog protein 4, ... | | Authors: | Centorrino, F, Wu, Q, Ottmann, C. | | Deposit date: | 2021-06-04 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A crystallography-based study of fragment extensions
into the 14-3-3 binding groove
To Be Published
|
|
8KAA
 
 | |
5CH7
 
 | | Crystal structure of the perchlorate reductase PcrAB - Phe164 gate switch intermediate - from Azospira suillum PS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ACETATE ION, ... | | Authors: | Tsai, C.-L, Tainer, J.A. | | Deposit date: | 2015-07-10 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Perchlorate Reductase Is Distinguished by Active Site Aromatic Gate Residues.
J.Biol.Chem., 291, 2016
|
|
7OYE
 
 | |
8SAA
 
 | |
8K8G
 
 | | De novo design protein -N9 | | Descriptor: | De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | De novo design protein -N9
To Be Published
|
|
7YDS
 
 | | The structure of the bispecific antibody targeted PD-L1 and 4-1BB | | Descriptor: | Anti-PDL1-VH-CH1, Anti-PDL1-VL-CL, Programmed cell death 1 ligand 1 | | Authors: | Gao, Y, Zhu, M, Liu, W.T, Cheng, L.S, Zhu, Z.L, Niu, L.W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A bispecific antibody targeted PD-L1 and 4-1BB induces a potent antitumor immune activity in colorectal cancer without systemic toxicity
To Be Published
|
|
7P0J
 
 | | Crystal structure of S.pombe Mdb1 BRCT domains | | Descriptor: | CITRIC ACID, DNA damage response protein Mdb1, MAGNESIUM ION, ... | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2021-06-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Phosphorylation-dependent assembly of DNA damage response systems and the central roles of TOPBP1.
DNA Repair (Amst), 108, 2021
|
|
8SBZ
 
 | |
5TON
 
 | | Crystal structure of AAT H143L mutant | | Descriptor: | Aspartate aminotransferase, cytoplasmic | | Authors: | Mueser, T.C, Dajnowicz, S, Kovalevsky, A. | | Deposit date: | 2016-10-18 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Direct evidence that an extended hydrogen-bonding network influences activation of pyridoxal 5'-phosphate in aspartate aminotransferase.
J. Biol. Chem., 292, 2017
|
|
8V8X
 
 | |
6MMZ
 
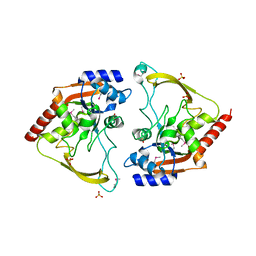 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, H29A mutant apoenzyme | | Descriptor: | Aminoglycoside N(3)-acetyltransferase, CHLORIDE ION, SULFATE ION | | Authors: | Stogios, P.J, Skarina, T, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
5BZ2
 
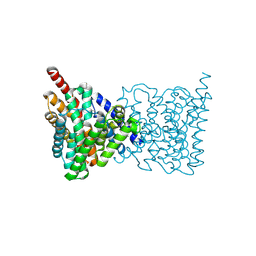 | |
5GIJ
 
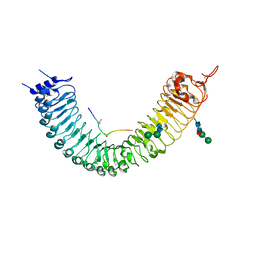 | | Crystal structure of TDR-TDIF complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich repeat receptor-like protein kinase TDR, ... | | Authors: | Morita, J, Kato, K, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2016-06-23 | | Release date: | 2016-08-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the plant receptor-like kinase TDR in complex with the TDIF peptide
Nat Commun, 7, 2016
|
|
6MNI
 
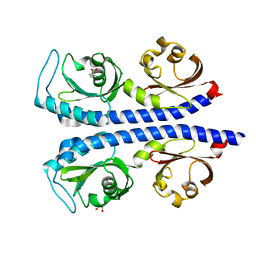 | | Structure of the tandem CACHE domain of PscC | | Descriptor: | CHLORIDE ION, GLYCEROL, Methyl-accepting chemotaxis protein, ... | | Authors: | Johnston, J.M, Gerth, M.L, Ehrhardt, M.K.G. | | Deposit date: | 2018-10-01 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structure of a double CACHE chemoreceptor ligand-binding domain from Pseudomonas syringae provides insights into the basis of proline recognition.
Biochem.Biophys.Res.Commun., 549, 2021
|
|
7OR7
 
 | | Ternary complex of 14-3-3 sigma, NotchpS1917 phosphopeptide, and WQ178 | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Centorrino, F, Wu, Q, Ottmann, C. | | Deposit date: | 2021-06-04 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A crystallography-based study of fragment extensions
into the 14-3-3 binding groove
To Be Published
|
|
5U5U
 
 | | CcP gateless cavity | | Descriptor: | ISONICOTINAMIDINE, PROTOPORPHYRIN IX CONTAINING FE, Peroxidase | | Authors: | Fischer, M, Shoichet, B.K. | | Deposit date: | 2016-12-07 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Testing inhomogeneous solvation theory in structure-based ligand discovery.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6MRV
 
 | | Sialidase26 co-crystallized with DANA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, Sialidase26 | | Authors: | Zaramela, L.S, Martino, C, Alisson-Silva, F, Rees, S.D, Diaz, S.L, Chuzel, L, Ganatra, M.B, Taron, C.H, Zuniga, C, Chang, G, Varki, A, Zengler, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates.
Nat Microbiol, 4, 2019
|
|
