7EW2
 
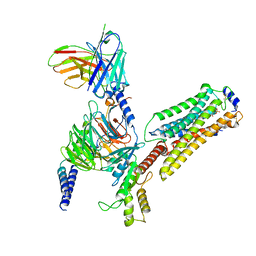 | | Cryo-EM structure of pFTY720-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | Descriptor: | (2~{S})-2-azanyl-4-(4-octylphenyl)-2-[[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxymethyl]butan-1-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
3DIK
 
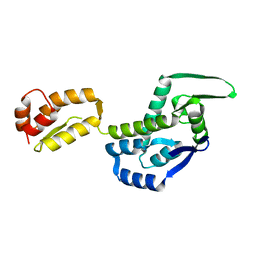 | |
1HN4
 
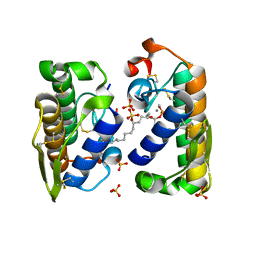 | | PROPHOSPHOLIPASE A2 DIMER COMPLEXED WITH MJ33, SULFATE, AND CALCIUM | | Descriptor: | 1-HEXADECYL-3-TRIFLUOROETHYL-SN-GLYCERO-2-PHOSPHATE METHANE, CALCIUM ION, PROPHOSPHOLIPASE A2, ... | | Authors: | Epstein, T.M, Pan, Y.H, Jain, M.K, Bahnson, B.J. | | Deposit date: | 2000-12-06 | | Release date: | 2001-12-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The basis for k(cat) impairment in prophospholipase A(2) from the anion-assisted dimer structure.
Biochemistry, 40, 2001
|
|
1EBG
 
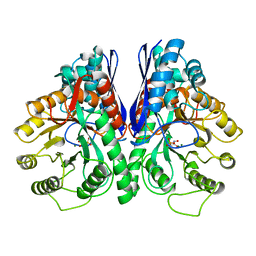 | |
3MLS
 
 | |
4MU7
 
 | | Crystal structure of cIAP1 BIR3 bound to T3450325 | | Descriptor: | (3S,10aS)-2-[(2S)-2-cyclohexyl-2-{[(2S)-2-(methylamino)butanoyl]amino}acetyl]-N-[(4R)-3,4-dihydro-2H-chromen-4-yl]-1,2,3,4,10,10a-hexahydropyrazino[1,2-a]indole-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Snell, G.P, Dougan, D.R. | | Deposit date: | 2013-09-20 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Design, synthesis, and biological activities of novel hexahydropyrazino[1,2-a]indole derivatives as potent inhibitors of apoptosis (IAP) proteins antagonists with improved membrane permeability across MDR1 expressing cells.
Bioorg.Med.Chem., 21, 2013
|
|
5NYN
 
 | | Crystal structure of the atypical poplar thioredoxin-like2.1 in complex with gluathione | | Descriptor: | GLUTATHIONE, SULFATE ION, Thioredoxin-like protein 2.1 | | Authors: | Chibani, K, Saul, F.A, Haouz, A, Rouhier, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-02-28 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural snapshots along the reaction mechanism of the atypical poplar thioredoxin-like2.1.
FEBS Lett., 592, 2018
|
|
7D3F
 
 | | Cryo-EM structure of human DUOX1-DUOXA1 in high-calcium state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Dual oxidase 1, ... | | Authors: | Chen, L, Wu, J.X. | | Deposit date: | 2020-09-19 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of human dual oxidase 1 complex in low-calcium and high-calcium states.
Nat Commun, 12, 2021
|
|
3MAD
 
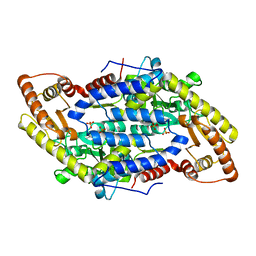 | |
3PXF
 
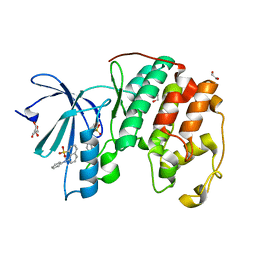 | | CDK2 in complex with two molecules of 8-anilino-1-naphthalene sulfonate | | Descriptor: | 1,2-ETHANEDIOL, 8-ANILINO-1-NAPHTHALENE SULFONATE, Cell division protein kinase 2 | | Authors: | Betzi, S, Alam, R, Schonbrunn, E. | | Deposit date: | 2010-12-09 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Potential Allosteric Ligand Binding Site in CDK2.
Acs Chem.Biol., 6, 2011
|
|
3M67
 
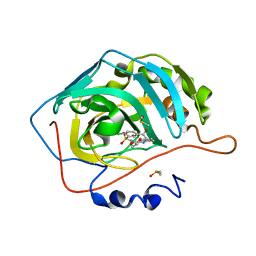 | | Crystal structure of human carbonic anhydrase isozyme II with 2-chloro-5-[(6,7-dihydro-1H-[1,4]dioxino[2,3-f]benzimidazol-2-ylsulfanyl)acetyl]benzenesulfonamide | | Descriptor: | 2-chloro-5-[(6,7-dihydro-1H-[1,4]dioxino[2,3-f]benzimidazol-2-ylsulfanyl)acetyl]benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Grazulis, S, Manakova, E, Golovenko, D. | | Deposit date: | 2010-03-15 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Indapamide-like benzenesulfonamides as inhibitors of carbonic anhydrases I, II, VII, and XIII
Bioorg.Med.Chem., 18, 2010
|
|
3ZXB
 
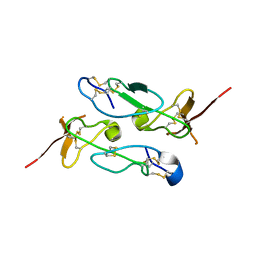 | |
6LXI
 
 | | Crystal structure of Z2B3 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1) | | Descriptor: | CALCIUM ION, Heavy chain of Z2B3 Fab, Light chain of Z2B3 Fab, ... | | Authors: | Jiang, H, Peng, W, Qi, J, Chai, Y, Song, H, Shi, Y, Gao, G.F, Wu, Y. | | Deposit date: | 2020-02-11 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Modification of an Anti-neuraminidase Human Antibody Restores Protection Efficacy against the Drifted Influenza Virus.
Mbio, 11, 2020
|
|
2PB2
 
 | | Structure of biosynthetic N-acetylornithine aminotransferase from Salmonella typhimurium: studies on substrate specificity and inhibitor binding | | Descriptor: | 1,2-ETHANEDIOL, Acetylornithine/succinyldiaminopimelate aminotransferase, GLYCEROL, ... | | Authors: | Rajaram, V, Ratna Prasuna, P, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2007-03-28 | | Release date: | 2007-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of biosynthetic N-acetylornithine aminotransferase from Salmonella typhimurium: Studies on substrate specificity and inhibitor binding
Proteins, 70, 2007
|
|
3AUJ
 
 | | Structure of diol dehydratase complexed with glycerol | | Descriptor: | CALCIUM ION, COBALAMIN, Diol dehydrase alpha subunit, ... | | Authors: | Yamanishi, M, Kinoshita, K, Fukuoka, M, Shibata, T, Tobimatsu, T, Toraya, T. | | Deposit date: | 2011-02-07 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Redesign of coenzyme B(12) dependent diol dehydratase to be resistant to the mechanism-based inactivation by glycerol and act on longer chain 1,2-diols
Febs J., 279, 2012
|
|
1VLP
 
 | |
3ZT0
 
 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | (4-CARBOXY-1,3-BENZODIOXOL-5-YL)-N-{2-[(4-METHOXYBENZYL)CARBAMOYL]BENZYL}-N-METHYLMETHANAMINIUM, INTEGRASE, SULFATE ION | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L, Deadman, J.J. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
2W5O
 
 | | Complex structure of the GH93 alpha-L-arabinofuranosidase of Fusarium graminearum with arabinobiose | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-L-ARABINOFURANOSIDASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Carapito, R, Imberty, A, Jeltsch, J.M, Byrns, S.C, Tam, P.H, Lowary, T.L, Varrot, A, Phalip, V. | | Deposit date: | 2008-12-11 | | Release date: | 2009-03-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular Basis of Arabinobio-Hydrolase Activity in Phytopathogenic Fungi. Crystal Structure and Catalytic Mechanism of Fusarium Graminearum Gh93 Exo-Alpha-L-Arabinanase.
J.Biol.Chem., 284, 2009
|
|
3ZSW
 
 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | (R)-[2-[[(2R)-butan-2-yl]carbamoyl]phenyl]methyl-[[(2S)-5-carboxy-2-(2-carboxyethyl)-2,3-dihydro-1,4-benzodioxin-6-yl]methyl]-prop-2-enyl-azanium, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L, Deadman, J.J. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
2NRU
 
 | | Crystal structure of IRAK-4 | | Descriptor: | 1-(3-HYDROXYPROPYL)-2-[(3-NITROBENZOYL)AMINO]-1H-BENZIMIDAZOL-5-YL PIVALATE, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Wang, Z, Liu, J, Walker, N.P.C. | | Deposit date: | 2006-11-02 | | Release date: | 2006-12-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of IRAK-4 kinase in complex with inhibitors: a serine/threonine kinase with tyrosine as a gatekeeper.
Structure, 14, 2006
|
|
3UQN
 
 | | Crystal structure of dihydrodipicolinate synthase from Acinetobacter baumannii complexed with Oxamic acid at 1.9 Angstrom resolution | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL, OXAMIC ACID | | Authors: | Singh, A, Kaushik, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-21 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of dihydrodipicolinate synthase from Acinetobacter baumannii complexed with Oxamic acid at 1.9 Angstrom resolution
To be Published
|
|
4IXV
 
 | | Crystal structure of human Arginase-2 complexed with inhibitor 2d: {(5R)-5-amino-5-carboxy-5-[1-(4-chlorobenzyl)piperidin-4-yl]pentyl}(trihydroxy)borate(1-) | | Descriptor: | Arginase-2, mitochondrial, BENZAMIDINE, ... | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Whitehouse, D, Beckett, P, Van Zandt, M.C, Ji, M.K, Ryder, T, Jagdmann, R, Andreoli, M, Olczak, J, Mazur, M, Czestkowski, W, Piotrowska, W, Schroeter, H, Golebiowski, A, Podjarny, A. | | Deposit date: | 2013-01-28 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis of quaternary alpha-amino acid-based arginase inhibitors via the Ugi reaction.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1HII
 
 | |
1DMS
 
 | | STRUCTURE OF DMSO REDUCTASE | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DMSO REDUCTASE, MOLYBDENUM (IV)OXIDE | | Authors: | Schneider, F, Loewe, J, Huber, R, Schindelin, H, Kisker, C, Knaeblein, J. | | Deposit date: | 1996-09-03 | | Release date: | 1998-07-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of dimethyl sulfoxide reductase from Rhodobacter capsulatus at 1.88 A resolution.
J.Mol.Biol., 263, 1996
|
|
4GYE
 
 | | MDR 769 HIV-1 Protease in Complex with Reduced P1F | | Descriptor: | P1F peptide, Protease | | Authors: | Dewdney, T.G, Wang, Y, Brunzelle, J, Reiter, S.J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2012-09-05 | | Release date: | 2013-10-30 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Ligand modifications to reduce the relative resistance of multi-drug resistant HIV-1 protease.
Bioorg.Med.Chem., 21, 2013
|
|
