5AVX
 
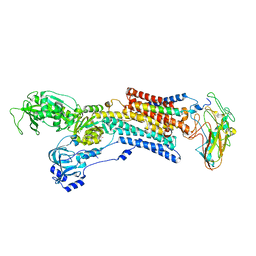 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 20 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
8R1D
 
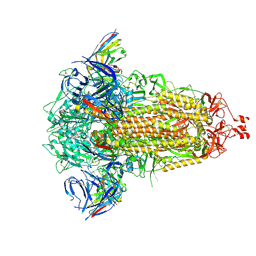 | | SD1-3 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SD1-3 Fab Heavy Chain, SD1-3 Fab Light Chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2023-11-01 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | The SARS-CoV-2 neutralizing antibody response to SD1 and its evasion by BA.2.86.
Nat Commun, 15, 2024
|
|
7ASN
 
 | |
7S2R
 
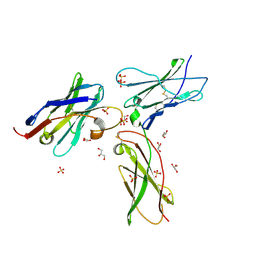 | | nanobody bound to IL-2Rg | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit gamma, ... | | Authors: | Glassman, C.R, Jude, K.M, Yen, M, Garcia, K.C. | | Deposit date: | 2021-09-03 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Facile discovery of surrogate cytokine agonists.
Cell, 185, 2022
|
|
8RX9
 
 | | LTA4 hydrolase in complex with compound3 | | Descriptor: | 1-[[5-[5-(1~{H}-pyrazol-5-yl)pyridin-2-yl]oxypyridin-2-yl]methyl]piperidin-4-ol, ACETATE ION, IMIDAZOLE, ... | | Authors: | Srinivas, H. | | Deposit date: | 2024-02-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Guided Elaboration of a Fragment-Like Hit into an Orally Efficacious Leukotriene A4 Hydrolase Inhibitor.
J.Med.Chem., 67, 2024
|
|
8RIY
 
 | | Human NUDT5 with ibrutinib derivative | | Descriptor: | 1-(1-methylpiperidin-4-yl)-3-(4-phenoxyphenyl)pyrazolo[3,4-d]pyrimidin-4-amine, ADP-sugar pyrophosphatase | | Authors: | Balikci-Akil, E, Elkins, J.M, Huber, K.V.M. | | Deposit date: | 2023-12-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Unexpected Noncovalent Off-Target Activity of Clinical BTK Inhibitors Leads to Discovery of a Dual NUDT5/14 Antagonist.
J.Med.Chem., 67, 2024
|
|
7ASM
 
 | |
7B74
 
 | | Chimeric Streptavidin With A Dimerization Domain For Artificial Transfer Hydrogenation | | Descriptor: | Streptavidin,Superoxide dismutase [Cu-Zn],Streptavidin, {N-(4-{[2-(amino-kappaN)ethyl]sulfamoyl-kappaN}phenyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}(chloro)[(1,2,3,4,5-eta)-1,2,3,4,5-pentamethylcyclopentadienyl]iridium(III) | | Authors: | Igareta, N.V, Ward, T.R. | | Deposit date: | 2020-12-09 | | Release date: | 2021-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Spiers Memorial Lecture: Shielding the active site: a streptavidin superoxide-dismutase chimera as a host protein for asymmetric transfer hydrogenation.
Faraday Disc.Chem.Soc, 2023
|
|
5EMN
 
 | | Crystal Structure of Human NADPH-Cytochrome P450 Reductase(A287P mutant) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Marohnic, C, Panda, S, Masters, B.S, Kim, J.J.K. | | Deposit date: | 2015-11-06 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Instability of the Human Cytochrome P450 Reductase A287P Variant Is the Major Contributor to Its Antley-Bixler Syndrome-like Phenotype.
J.Biol.Chem., 291, 2016
|
|
5AYN
 
 | | Crystal structure of a bacterial homologue of iron transporter ferroportin in outward-facing state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, POTASSIUM ION, Solute carrier family 39 (Iron-regulated transporter) | | Authors: | Taniguchi, R, Kato, H.E, Font, J, Deshpande, C.N, Ishitani, R, Jormakka, M, Nureki, O. | | Deposit date: | 2015-08-25 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Outward- and inward-facing structures of a putative bacterial transition-metal transporter with homology to ferroportin
Nat Commun, 6, 2015
|
|
7AU6
 
 | |
7ATN
 
 | |
7S4K
 
 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.34 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
7ATE
 
 | | Cytochrome c oxidase structure in P-state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, ... | | Authors: | Kolbe, F, Safarian, S, Michel, H. | | Deposit date: | 2020-10-30 | | Release date: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cytochrome c oxidase structure in P-state
To Be Published
|
|
8R8T
 
 | |
7AU3
 
 | | Cytochrome c oxidase structure in F-state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Kolbe, F, Safarian, S, Michel, H. | | Deposit date: | 2020-11-02 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Cytochrome c oxidase structure in F-state
To Be Published
|
|
8RQQ
 
 | | In meso structure of the adenosine A2a G protein-coupled receptor, A2aR, in 7.10 monoacylglycerol | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, 7.10 monoacylglycerol (R-form), 7.10 monoacylglycerol (S-form), ... | | Authors: | Smithers, L, Krawinski, P, Caffrey, M. | | Deposit date: | 2024-01-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | 7.10 MAG. A Novel Host Monoacylglyceride for In Meso (Lipid Cubic Phase) Crystallization of Membrane Proteins.
Cryst.Growth Des., 24, 2024
|
|
7S0S
 
 | | M. tuberculosis ribosomal RNA methyltransferase TlyA bound to M. smegmatis 50S ribosomal subunit | | Descriptor: | 16S/23S rRNA (Cytidine-2'-O)-methyltransferase TlyA, 23S rRNA, 50S ribosomal protein L10, ... | | Authors: | Laughlin, Z.T, Dunham, C.M, Conn, G.L. | | Deposit date: | 2021-08-31 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | 50S subunit recognition and modification by the Mycobacterium tuberculosis ribosomal RNA methyltransferase TlyA.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5C0X
 
 | |
5U1D
 
 | |
8S5B
 
 | | Crystal structure of the sulfoquinovosyl binding protein (SmoF) from A. tumefaciens sulfo-SMO pathway in complex with SQOctyl ligand | | Descriptor: | Sulfoquinovosyl glycerol-binding protein SmoF, [(2~{S},3~{S},4~{S},5~{R},6~{S})-6-octoxy-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Snow, A.J.D, Sharma, M, Davies, G.J. | | Deposit date: | 2024-02-23 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Capture-and-release of a sulfoquinovose-binding protein on sulfoquinovose-modified agarose.
Org.Biomol.Chem., 22, 2024
|
|
6PZB
 
 | |
3OR6
 
 | | On the structural basis of modal gating behavior in K+channels - E71Q | | Descriptor: | POTASSIUM ION, Voltage-gated potassium channel, antibody fab fragment heavy chain, ... | | Authors: | Chakrapani, S, Cordero-Morales, J.F, Jogini, V, Perozo, E. | | Deposit date: | 2010-09-06 | | Release date: | 2011-01-05 | | Last modified: | 2018-08-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | On the structural basis of modal gating behavior in K(+) channels.
Nat.Struct.Mol.Biol., 18, 2011
|
|
5EHC
 
 | | Co-crystal structure of eIF4E with nucleotide mimetic inhibitor. | | Descriptor: | 3-[[(2~{R},3~{S},4~{R},5~{R})-5-[2-azanyl-7-[(3-chlorophenyl)methyl]-6-oxidanylidene-1~{H}-purin-7-ium-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylamino]-4-oxidanyl-cyclobut-3-ene-1,2-dione, Eukaryotic translation initiation factor 4 gamma 1, Eukaryotic translation initiation factor 4E | | Authors: | Nowicki, M.W, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2015-10-28 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design of nucleotide-mimetic and non-nucleotide inhibitors of the translation initiation factor eIF4E: Synthesis, structural and functional characterisation.
Eur.J.Med.Chem., 124, 2016
|
|
5C3F
 
 | | Crystal structure of Mcl-1 bound to BID-MM | | Descriptor: | BID-MM, GLYCEROL, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Miles, J.A, Yeo, D.J, Rowell, P, Rodriguez-Marin, S, Pask, C.M, Warriner, S.L, Edwards, T.A, Wilson, A.J. | | Deposit date: | 2015-06-17 | | Release date: | 2016-04-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Hydrocarbon constrained peptides - understanding preorganisation and binding affinity.
Chem Sci, 7, 2016
|
|
