4V6H
 
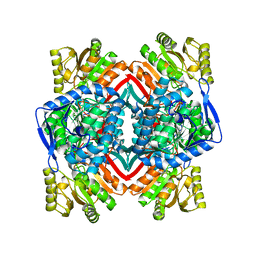 | |
4V7L
 
 | | The structures of viomycin bound to the 70S ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Stanley, R.E, Blaha, G. | | Deposit date: | 2009-11-12 | | Release date: | 2014-07-09 | | Last modified: | 2018-07-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4V4Z
 
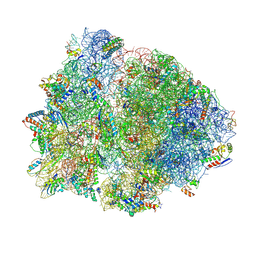 | | 70S Thermus thermophilous ribosome functional complex with mRNA and E- and P-site tRNAs at 4.5A. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Jenner, L, Yusupova, G, Rees, B, Moras, D, Yusupov, M. | | Deposit date: | 2006-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (4.51 Å) | | Cite: | Structural basis for messenger RNA movement on the ribosome.
Nature, 444, 2006
|
|
4V9R
 
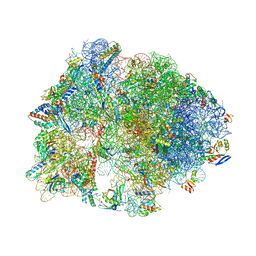 | | Crystal structure of antibiotic DITYROMYCIN bound to 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
2FVU
 
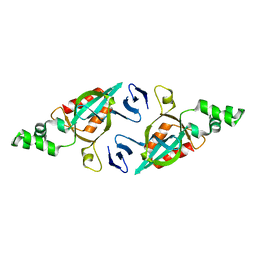 | | Structure of the yeast Sir3 BAH domain | | Descriptor: | Regulatory protein SIR3 | | Authors: | Xu, R.M. | | Deposit date: | 2006-01-31 | | Release date: | 2006-09-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of the Saccharomyces cerevisiae Sir3 BAH domain.
Mol.Cell.Biol., 26, 2006
|
|
2FPY
 
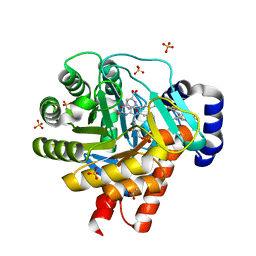 | | Dual binding mode of a novel series of DHODH inhibitors | | Descriptor: | 3-({[3,5-DIFLUORO-3'-(TRIFLUOROMETHOXY)BIPHENYL-4-YL]AMINO}CARBONYL)THIOPHENE-2-CARBOXYLIC ACID, ACETATE ION, Dihydroorotate dehydrogenase, ... | | Authors: | Baumgartner, R, Leban, J. | | Deposit date: | 2006-01-17 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dual binding mode of a novel series of DHODH inhibitors.
J.Med.Chem., 49, 2006
|
|
2G3J
 
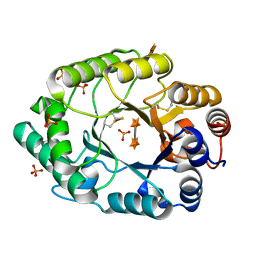 | | Structure of S.olivaceoviridis xylanase Q88A/R275A mutant | | Descriptor: | PHOSPHATE ION, Xylanase, alpha-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Diertavitian, S, Kaneko, S, Fujimoto, Z, Kuno, A, Johansson, E, Lo Leggio, L. | | Deposit date: | 2006-02-20 | | Release date: | 2007-03-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based engineering of glucose specificity in a family 10 xylanase from Streptomyces olivaceoviridis E-86
PROCESS BIOCHEM, 47, 2012
|
|
2FRX
 
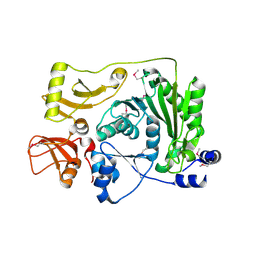 | | Crystal structure of YebU, a m5C RNA methyltransferase from E.coli | | Descriptor: | Hypothetical protein yebU | | Authors: | Erlandsen, H, Nordlund, P, Hallberg, B.M, Johnson, K.A, Ericsson, U.B. | | Deposit date: | 2006-01-20 | | Release date: | 2006-08-29 | | Last modified: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of the RNA m5C methyltransferase YebU from Escherichia coli reveals a C-terminal RNA-recruiting PUA domain
J.Mol.Biol., 360, 2006
|
|
2FQI
 
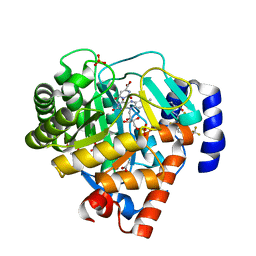 | | dual binding modes of a novel series of DHODH inhibitors | | Descriptor: | 2-({[2,3,5,6-TETRAFLUORO-3'-(TRIFLUOROMETHOXY)BIPHENYL-4-YL]AMINO}CARBONYL)CYCLOPENTA-1,3-DIENE-1-CARBOXYLIC ACID, ACETATE ION, Dihydroorotate dehydrogenase, ... | | Authors: | Baumgartner, R, Leban, J. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dual binding mode of a novel series of DHODH inhibitors.
J.Med.Chem., 49, 2006
|
|
2FPZ
 
 | | Human tryptase with 2-amino benzimidazole | | Descriptor: | 2H-BENZOIMIDAZOL-2-YLAMINE, Tryptase beta-2 | | Authors: | Somoza, J.R. | | Deposit date: | 2006-01-17 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided design of Peptide-based tryptase inhibitors.
Biochemistry, 45, 2006
|
|
2FSZ
 
 | |
2FS4
 
 | | Ketopiperazine-Based Renin Inhibitors: Optimization of the C ring | | Descriptor: | (6R)-6-({[1-(3-HYDROXYPROPYL)-1,7-DIHYDROQUINOLIN-7-YL]OXY}METHYL)-1-(4-{3-[(2-METHOXYBENZYL)OXY]PROPOXY}PHENYL)PIPERAZIN-2-ONE, Renin | | Authors: | Holsworth, D.D, Cai, C, Cheng, X.-M, Cody, W.L, Downing, D.M, Erasga, N, Lee, C, Powell, N.A, Edmunds, J.J, Stier, M, Jalaie, M, Zhang, E, McConnell, P, Ryan, M.J, Bryant, J, Li, T, Kasani, A, Hall, E, Subedi, R, Rahim, M, Maiti, S. | | Deposit date: | 2006-01-20 | | Release date: | 2006-06-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ketopiperazine-Based Renin Inhibitors: Optimization of the "C" Ring
BIOORG.MED.CHEM.LETT., 16, 2006
|
|
2FVV
 
 | | Human Diphosphoinositol polyphosphate phosphohydrolase 1 | | Descriptor: | CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Hallberg, B.M, Kursula, P, Ogg, D, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Flodin, S, Graslund, S, Hammarstrom, M, Hogbom, M, Holmberg-Schiavone, L, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, van den Berg, S, Weigelt, J, Persson, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-31 | | Release date: | 2006-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of human diphosphoinositol phosphatase 1
Proteins, 77, 2009
|
|
2G0D
 
 | | Nisin cyclase | | Descriptor: | Nisin biosynthesis protein nisC, ZINC ION | | Authors: | Nair, S.K. | | Deposit date: | 2006-02-12 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure and mechanism of the lantibiotic cyclase involved in nisin biosynthesis
Science, 311, 2006
|
|
2G3B
 
 | | Crystal structure of putative TetR-family transcriptional regulator from Rhodococcus sp. | | Descriptor: | GLYCEROL, putative TetR-family transcriptional regulator | | Authors: | Chruszcz, M, Evdokimova, E, Cymborowski, M, Kagan, O, Wang, S, Koclega, K.D, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-17 | | Release date: | 2006-03-14 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative TetR-family transcriptional regulator from Rhodococcus sp.
To be Published
|
|
2G4F
 
 | | Structure of S.olivaceoviridis xylanase Q88A/R275A mutant | | Descriptor: | Hydrolase | | Authors: | Diertavitian, S, Kaneko, S, Fujimoto, Z, Kuno, A, Johansson, E, Lo Leggio, L. | | Deposit date: | 2006-02-22 | | Release date: | 2007-03-06 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-based engineering of glucose specificity in a family 10 xylanase from Streptomyces olivaceoviridis E-86
PROCESS BIOCHEM, 47, 2012
|
|
2G5F
 
 | | The structure of MbtI from Mycobacterium Tuberculosis, the first enzyme in the synthesis of Mycobactin, reveals it to be a salicylate synthase | | Descriptor: | COG0147: Anthranilate/para-aminobenzoate synthases component I, GLYCEROL, IMIDAZOLE, ... | | Authors: | Harrison, A.J, Lott, J.S, Yu, M, Ramsay, R, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-02-22 | | Release date: | 2006-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of MbtI from Mycobacterium tuberculosis, the First Enzyme in the Biosynthesis of the Siderophore Mycobactin, Reveals It To Be a Salicylate Synthase
J.Bacteriol., 188, 2006
|
|
2GDV
 
 | |
2GG2
 
 | | Novel bacterial methionine aminopeptidase inhibitors | | Descriptor: | 5-IMINO-4-(3-TRIFLUOROMETHYL-PHENYLAZO)-5H-PYRAZOL-3-YLAMINE, COBALT (II) ION, Methionine aminopeptidase, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Serendipitous discovery of novel bacterial methionine aminopeptidase inhibitors.
Proteins, 66, 2007
|
|
2GG0
 
 | | Novel bacterial methionine aminopeptidase inhibitors | | Descriptor: | COBALT (II) ION, METHYL N-{(2S,3R)-3-AMINO-2-HYDROXY-3-[4-(TRIFLUOROMETHYL)PHENYL]PROPANOYL}ALANYLGLYCINATE, Methionine aminopeptidase, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Serendipitous discovery of novel bacterial methionine aminopeptidase inhibitors.
Proteins, 66, 2007
|
|
2GGB
 
 | | Novel bacterial methionine aminopeptidase inhibitors | | Descriptor: | COBALT (II) ION, METHYL N-[(2S,3R)-3-AMINO-2-HYDROXYHEPTANOYL]-L-SERYL-L-LEUCINATE, Methionine aminopeptidase, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Serendipitous discovery of novel bacterial methionine aminopeptidase inhibitors.
Proteins, 66, 2007
|
|
2FVZ
 
 | | Human Inositol Monophosphosphatase 2 | | Descriptor: | Inositol monophosphatase 2 | | Authors: | Ogg, D, Hallberg, B.M, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Graslund, S, Hammarstrom, M, Hogbom, M, Holmberg-Schiavone, L, Kotenyova, T, Kursula, P, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Van Den Berg, S, Weigelt, J, Thorsell, A.G, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-31 | | Release date: | 2006-02-21 | | Last modified: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Human Inositol Monophosphatase 2
To be published
|
|
2G82
 
 | |
2GAF
 
 | | Crystal Structure of the Vaccinia Polyadenylate Polymerase Heterodimer (apo form) | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, Poly(A) polymerase catalytic subunit | | Authors: | Moure, C.M, Bowman, B.R, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 2006-03-08 | | Release date: | 2006-05-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the vaccinia virus polyadenylate polymerase heterodimer: insights into ATP selectivity and processivity.
Mol.Cell, 22, 2006
|
|
2G6X
 
 | |
