5WFJ
 
 | | THE JAK3 KINASE DOMAIN IN COMPLEX WITH A COVALENT INHIBITOR | | Descriptor: | 4-({[3-(propanoylamino)phenyl]methyl}amino)pyrrolo[1,2-b]pyridazine-3-carboxamide, Tyrosine-protein kinase JAK3 | | Authors: | Sack, J. | | Deposit date: | 2017-07-12 | | Release date: | 2017-10-04 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of highly potent, selective, covalent inhibitors of JAK3.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
8BZY
 
 | | Structure of SLC40/ferroportin in complex with vamifeport and synthetic nanobody Sy3 in occluded conformation | | Descriptor: | 2-[2-[2-(1~{H}-benzimidazol-2-yl)ethylamino]ethyl]-~{N}-[(3-fluoranylpyridin-2-yl)methyl]-1,3-oxazole-4-carboxamide, DIUNDECYL PHOSPHATIDYL CHOLINE, Solute carrier family 40 member 1, ... | | Authors: | Lehmann, E.F, Liziczai, M, Drozdzyk, K, Dutzler, R, Manatschal, C. | | Deposit date: | 2022-12-15 | | Release date: | 2023-03-22 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structures of ferroportin in complex with its specific inhibitor vamifeport.
Elife, 12, 2023
|
|
8C02
 
 | | Structure of SLC40/ferroportin in complex with synthetic nanobody Sy3 in occluded conformation | | Descriptor: | Solute carrier family 40 member 1, Sybody3 | | Authors: | Lehmann, E.F, Liziczai, M, Drozdzyk, K, Dutzler, R, Manatschal, C. | | Deposit date: | 2022-12-15 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (4.09 Å) | | Cite: | Structures of ferroportin in complex with its specific inhibitor vamifeport.
Elife, 12, 2023
|
|
8COA
 
 | | in situ Subtomogram average of Immature Rotavirus TLP spike | | Descriptor: | Intermediate capsid protein VP6, Outer capsid glycoprotein VP7, Outer capsid protein VP4 | | Authors: | Shah, P.N.M, Stuart, D.I. | | Deposit date: | 2023-02-27 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Characterization of the rotavirus assembly pathway in situ using cryoelectron tomography.
Cell Host Microbe, 31, 2023
|
|
3LAK
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with N1-heterocycle pyrimidinedione non-nucleoside inhibitor | | Descriptor: | 3-({3-[(2-amino-6-fluoropyridin-4-yl)methyl]-5-(1-methylethyl)-2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl}carbonyl)-5-methylbenzonitrile, CHLORIDE ION, HIV Reverse transcriptase, ... | | Authors: | Lansdon, E.B, Mitchell, M.L. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | N1-Heterocyclic pyrimidinediones as non-nucleoside inhibitors of HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4Z1Z
 
 | |
4Z20
 
 | |
5WJI
 
 | | Crystal structure of the F61S mutant of HsNUDT16 | | Descriptor: | ACETIC ACID, CHLORIDE ION, SULFATE ION, ... | | Authors: | Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2017-07-23 | | Release date: | 2018-10-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analyses of NudT16-ADP-ribose complexes direct rational design of mutants with improved processing of poly(ADP-ribosyl)ated proteins.
Sci Rep, 9, 2019
|
|
5WJZ
 
 | | Cryo-EM structure of B. subtilis flagellar filaments E115G | | Descriptor: | Flagellin | | Authors: | Wang, F, Burrage, A.M, Orlova, A, Kearns, D.B, Egelman, E.H. | | Deposit date: | 2017-07-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | A structural model of flagellar filament switching across multiple bacterial species.
Nat Commun, 8, 2017
|
|
3LJ3
 
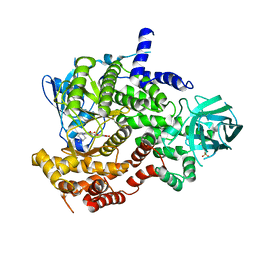 | | PI3-kinase-gamma with a pyrrolopyridine-benzofuran inhibitor | | Descriptor: | (2Z)-4,6-dihydroxy-2-{[1-methyl-4-(4-methylpiperazin-1-yl)-1H-pyrrolo[2,3-b]pyridin-3-yl]methylidene}-1-benzofuran-3(2H)-one, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Bard, J, Svenson, K. | | Deposit date: | 2010-01-25 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery and optimization of 2-(4-substituted-pyrrolo[2,3-b]pyridin-3-yl)methylene-4-hydroxybenzofuran-3(2H)-ones as potent and selective ATP-competitive inhibitors of the mammalian target of rapamycin (mTOR).
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4YZU
 
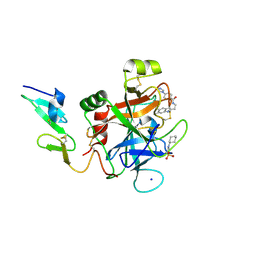 | |
8CDK
 
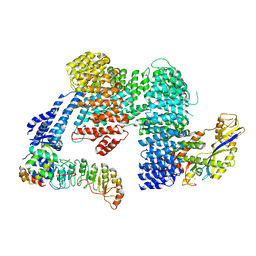 | |
5WNR
 
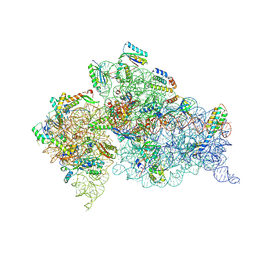 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S Ribosomal RNA rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | DeMirci, H. | | Deposit date: | 2017-08-01 | | Release date: | 2018-02-21 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | 2'-O-methylation in mRNA disrupts tRNA decoding during translation elongation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8CDJ
 
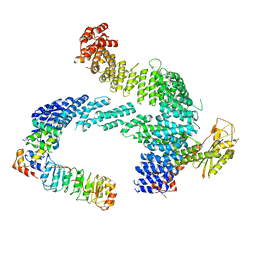 | | CAND1 b-hairpin++-SCF-SKP2 CAND1 rolling SCF engaged | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2023-01-31 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
1NZY
 
 | |
5W8A
 
 | |
3HVK
 
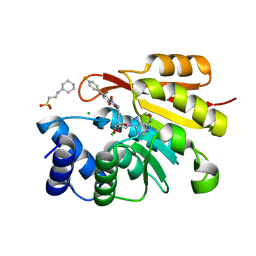 | | Rat catechol O-methyltransferase in complex with a catechol-type, purine-containing bisubstrate inhibitor - humanized form | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2009-06-16 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular recognition at the active site of catechol-o-methyltransferase: energetically favorable replacement of a water molecule imported by a bisubstrate inhibitor.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
2XFG
 
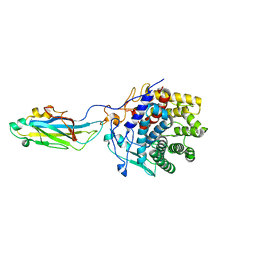 | | Reassembly and co-crystallization of a family 9 processive endoglucanase from separately expressed GH9 and CBM3c modules | | Descriptor: | CALCIUM ION, CHLORIDE ION, ENDOGLUCANASE 1 | | Authors: | Petkun, S, Lamed, R, Jindou, S, Burstein, T, Yaniv, O, Shoham, Y, Shimon, J.W.L, Bayer, E.A, Frolow, F. | | Deposit date: | 2010-05-24 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | Reassembly and Co-Crystallization of a Family 9 Processive Endoglucanase from its Component Parts: Structural and Functional Significance of Intermodular Linker
Peerj, 3, 2015
|
|
2Y25
 
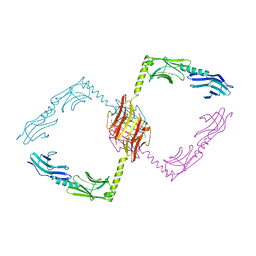 | |
2XUU
 
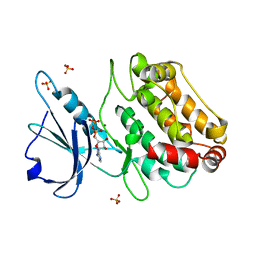 | | Crystal structure of a DAP-kinase 1 mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DEATH-ASSOCIATED PROTEIN KINASE 1, MAGNESIUM ION, ... | | Authors: | de Diego, I, Kuper, J, Lehmann, F, Wilmanns, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Pef/Y Substrate Recognition and Signature Motif Plays a Critical Role in Dapk-Related Kinase Activity.
Chem.Biol., 21, 2014
|
|
3HWK
 
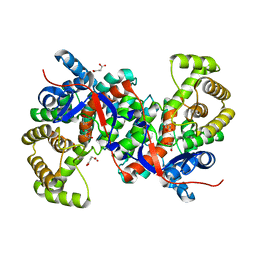 | |
1Y6X
 
 | |
5ZEC
 
 | | Crystal structure of Kluyveromyces polyspora ADH (KpADH) mutant (Q136N/F161V/S196G/E214G/S237C) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ETHANOL, ... | | Authors: | Wang, Y, ZHou, J.Y, Hou, X.D, Xu, G.C, Rao, Y.J, Wu, L, Zhou, J.H, Ni, Y. | | Deposit date: | 2018-02-27 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Structural Insight into Enantioselective Inversion of an Alcohol Dehydrogenase Reveals a "Polar Gate" in Stereorecognition of Diaryl Ketones.
J. Am. Chem. Soc., 140, 2018
|
|
7Y70
 
 | | RLGSGG-AtPRT6 UBR box (P4332) | | Descriptor: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | Authors: | Kim, L, Song, H.K. | | Deposit date: | 2022-06-21 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7XWD
 
 | | Apo-AtPRT6 UBR box | | Descriptor: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | Authors: | Kim, L, Song, H.K. | | Deposit date: | 2022-05-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
