2HM4
 
 | | Nematocyst Outer Wall Antigen, NW1 K21P | | Descriptor: | Nematocyst outer wall antigen | | Authors: | Meier, S, Jensen, P.R, Grzesiek, S, Oezbek, S. | | Deposit date: | 2006-07-11 | | Release date: | 2007-02-06 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Continuous molecular evolution of protein-domain structures by single amino Acid changes.
Curr.Biol., 17, 2007
|
|
2HP8
 
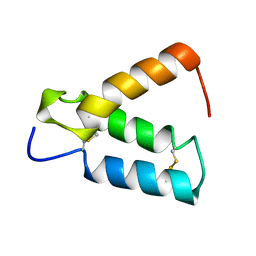 | | SOLUTION STRUCTURE OF HUMAN P8-MTCP1, A CYSTEINE-RICH PROTEIN ENCODED BY THE MTCP1 ONCOGENE,REVEALS A NEW ALPHA-HELICAL ASSEMBLY MOTIF, NMR, 30 STRUCTURES | | Descriptor: | Cx9C motif-containing protein 4 | | Authors: | Barthe, P, Chiche, L, Strub, M.P, Roumestand, C. | | Deposit date: | 1997-08-26 | | Release date: | 1998-03-04 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human p8MTCP1, a cysteine-rich protein encoded by the MTCP1 oncogene, reveals a new alpha-helical assembly motif.
J.Mol.Biol., 274, 1997
|
|
2HM6
 
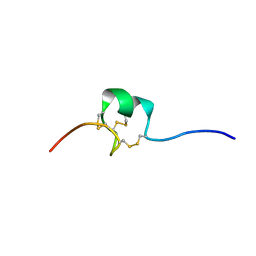 | | Nematocyst outer wall antigen, NW1 G11V K21P | | Descriptor: | Nematocyst outer wall antigen | | Authors: | Meier, S, Jensen, P.R, Grzesiek, S, Oezbek, S. | | Deposit date: | 2006-07-11 | | Release date: | 2007-02-06 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Continuous molecular evolution of protein-domain structures by single amino Acid changes.
Curr.Biol., 17, 2007
|
|
5JPI
 
 | | 2.15 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with D-Eritadenine and NAD | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Adenosylhomocysteinase, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-03 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.15 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with D-Eritadenine and NAD.
To Be Published
|
|
5JXW
 
 | | 2.25 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Neplanocin-A and NAD | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Adenosylhomocysteinase, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-13 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2.25 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Neplanocin-A and NAD
To Be Published
|
|
8K3S
 
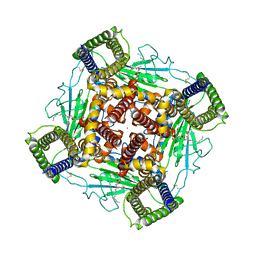 | | Structure of PKD2-F604P complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Chen, M.Y, Su, Q, Wang, Z.F, Yu, Y. | | Deposit date: | 2023-07-16 | | Release date: | 2024-04-03 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular and structural basis of the dual regulation of the polycystin-2 ion channel by small-molecule ligands.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3BBA
 
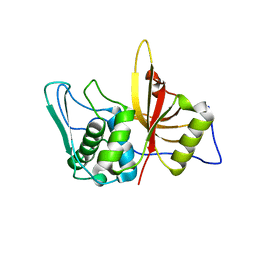 | | Structure of active wild-type Prevotella intermedia interpain A cysteine protease | | Descriptor: | interpain A | | Authors: | Mallorqui-Fernandez, N, Manandhar, S.P, Mallorqui-Fernandez, G, Uson, I, Wawrzonek, K, Kantyka, T, Sola, M, Thogersen, I.B, Enghild, J.J, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2007-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A New Autocatalytic Activation Mechanism for Cysteine Proteases Revealed by Prevotella intermedia Interpain A
J.Biol.Chem., 283, 2008
|
|
3B8B
 
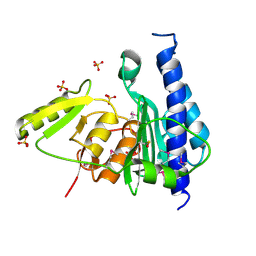 | | Crystal structure of CysQ from Bacteroides thetaiotaomicron, a bacterial member of the inositol monophosphatase family | | Descriptor: | CHLORIDE ION, CysQ, sulfite synthesis pathway protein, ... | | Authors: | Cuff, M.E, Mulligan, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-10-31 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of CysQ from Bacteroides thetaiotaomicron, a bacterial member of the inositol monophosphatase family.
TO BE PUBLISHED
|
|
7TUS
 
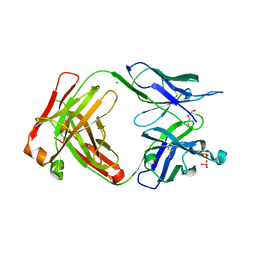 | |
4A2N
 
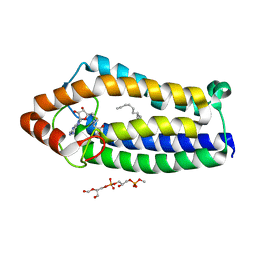 | | Crystal Structure of Ma-ICMT | | Descriptor: | CARDIOLIPIN, ISOPRENYLCYSTEINE CARBOXYL METHYLTRANSFERASE, PALMITIC ACID, ... | | Authors: | Yang, J, Kulkarni, K, Manolaridis, I, Zhang, Z, Dodd, R.B, Mas-Droux, C, Barford, D. | | Deposit date: | 2011-09-27 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mechanism of Isoprenylcysteine Carboxyl Methylation from the Crystal Structure of the Integral Membrane Methyltransferase Icmt.
Mol.Cell, 44, 2011
|
|
9IXD
 
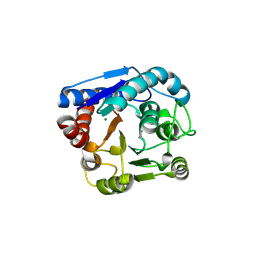 | |
1EXK
 
 | | SOLUTION STRUCTURE OF THE CYSTEINE-RICH DOMAIN OF THE ESCHERICHIA COLI CHAPERONE PROTEIN DNAJ. | | Descriptor: | DNAJ PROTEIN, ZINC ION | | Authors: | Martinez-Yamout, M, Legge, G.B, Zhang, O, Wright, P.E, Dyson, H.J. | | Deposit date: | 2000-05-03 | | Release date: | 2000-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cysteine-rich domain of the Escherichia coli chaperone protein DnaJ.
J.Mol.Biol., 300, 2000
|
|
6DU8
 
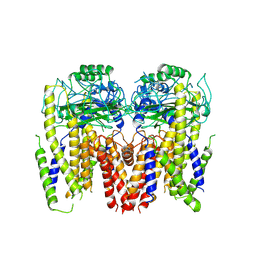 | | Human Polycsytin 2-l1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Polycystic kidney disease 2-like 1 protein | | Authors: | Hulse, R.E, Clapham, D.E, Li, Z, Huang, R.K, Zhang, J. | | Deposit date: | 2018-06-20 | | Release date: | 2018-07-25 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Cryo-EM structure of the polycystin 2-l1 ion channel.
Elife, 7, 2018
|
|
3AAX
 
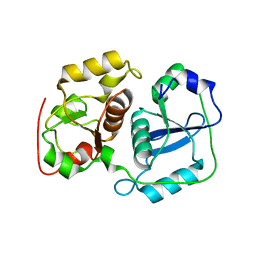 | | Crystal structure of probable thiosulfate sulfurtransferase cysa3 (RV3117) from Mycobacterium tuberculosis: monoclinic FORM | | Descriptor: | Putative thiosulfate sulfurtransferase | | Authors: | Sankaranarayanan, R, Witholt, S.J, Cherney, M.M, Garen, C.R, Cherney, L.T, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of probable thiosulfate sulfurtransferase CysA3 (Rv3117) from Mycobacterium tuberculosis
To be Published
|
|
3AAY
 
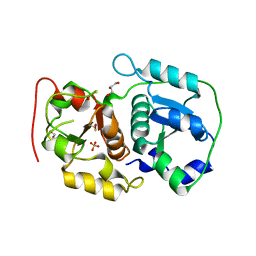 | | Crystal structure of probable thiosulfate sulfurtransferase CYSA3 (RV3117) from Mycobacterium tuberculosis: orthorhombic form | | Descriptor: | GLYCEROL, Putative thiosulfate sulfurtransferase, SULFATE ION | | Authors: | Sankaranarayanan, R, Witholt, S.J, Cherney, M.M, Garen, C.R, Cherney, L.T, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of probable thiosulfate sulfurtransferase CysA3 (Rv3117) from Mycobacterium tuberculosis
To be Published
|
|
2NNR
 
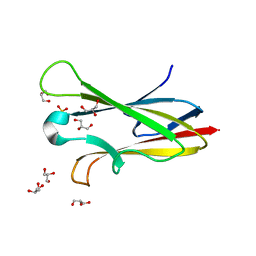 | | Crystal structure of chagasin, cysteine protease inhibitor from Trypanosoma cruzi | | Descriptor: | CHLORIDE ION, Chagasin, GLYCEROL, ... | | Authors: | Redzynia, I, Bujacz, G, Ljunggren, A, Jaskolski, M, Abrahamson, M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the parasite protease inhibitor chagasin in complex with a host target cysteine protease
J.Mol.Biol., 371, 2007
|
|
3DHC
 
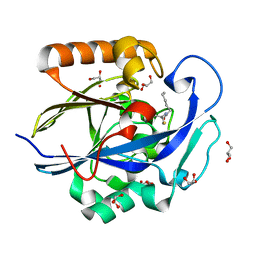 | | 1.3 Angstrom Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homocysteine Bound to The catalytic Metal Center | | Descriptor: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homocysteine, ... | | Authors: | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
8PB3
 
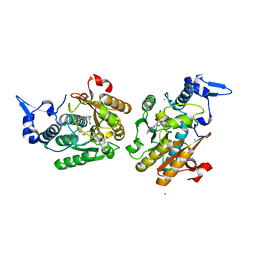 | | PsiM in complex with SAH and norbaeocystin, monoclinic crystal form | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Norbaeocystin, ... | | Authors: | Werten, S, Hudspeth, J, Rupp, B. | | Deposit date: | 2023-06-08 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Methyl transfer in psilocybin biosynthesis.
Nat Commun, 15, 2024
|
|
8PB4
 
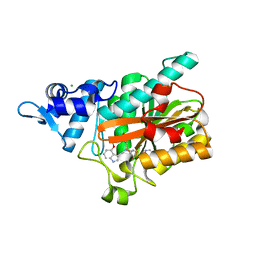 | | PsiM in complex with SAH and norbaeocystin, orthorhombic crystal form | | Descriptor: | CHLORIDE ION, Norbaeocystin, Psilocybin synthase, ... | | Authors: | Werten, S, Hudspeth, J, Rupp, B. | | Deposit date: | 2023-06-08 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Methyl transfer in psilocybin biosynthesis.
Nat Commun, 15, 2024
|
|
5UTU
 
 | | 2.65 Angstrom Resolution Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with SAH and NAD | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Stam, J, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 2.65 Angstrom Resolution Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with SAH and NAD
To Be Published
|
|
3L5L
 
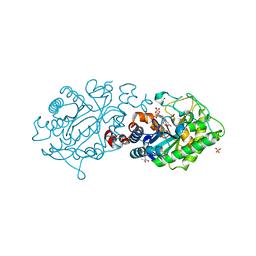 | | Xenobiotic Reductase A - oxidized | | Descriptor: | (R,R)-2,3-BUTANEDIOL, FLAVIN MONONUCLEOTIDE, SULFATE ION, ... | | Authors: | Spiegelhauer, O, Dobbek, H. | | Deposit date: | 2009-12-22 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Cysteine as a modulator residue in the active site of xenobiotic reductase A: a structural, thermodynamic and kinetic study
J.Mol.Biol., 398, 2010
|
|
5HM8
 
 | | 2.85 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Adenosine and NAD. | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Halavaty, A, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-15 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | 2.85 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Adenosine and NAD.
To Be Published
|
|
3L68
 
 | | Xenobiotic Reductase A - C25S variant with coumarin | | Descriptor: | (R,R)-2,3-BUTANEDIOL, COUMARIN, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Spiegelhauer, O, Dobbek, H. | | Deposit date: | 2009-12-23 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cysteine as a modulator residue in the active site of xenobiotic reductase A: a structural, thermodynamic and kinetic study
J.Mol.Biol., 398, 2010
|
|
3DU1
 
 | |
3L66
 
 | | Xenobiotic Reductase A - C25A Variant with Coumarin | | Descriptor: | (R,R)-2,3-BUTANEDIOL, COUMARIN, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Spiegelhauer, O, Dobbek, H. | | Deposit date: | 2009-12-23 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Cysteine as a modulator residue in the active site of xenobiotic reductase A: a structural, thermodynamic and kinetic study
J.Mol.Biol., 398, 2010
|
|
