8K5U
 
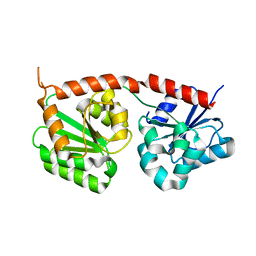 | | Se-glycosyltransferase RsSenB | | Descriptor: | Se glycosyltransferase | | Authors: | Huang, W, Long, F. | | Deposit date: | 2023-07-24 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of RsSenB at 2.15 Angstroms resolution
To Be Published
|
|
7TOA
 
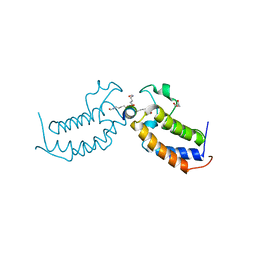 | |
7P9R
 
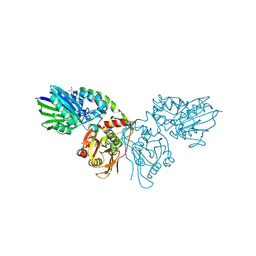 | | Crystal structure of CD73 in complex with GMP in the open form | | Descriptor: | 5'-nucleotidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-27 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
5FSE
 
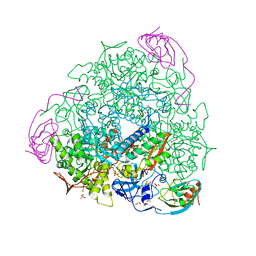 | | 2.07 A resolution 1,4-Benzoquinone inhibited Sporosarcina pasteurii urease | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Musiani, F, Ciurli, S. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Inactivation of Urease by 1,4-Benzoquinone: Chemistry at the Protein Surface.
Dalton Trans, 45, 2016
|
|
7TH5
 
 | | Thermus thermophilus methylenetetrahydrofolate reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Methylenetetrahydrofolate reductase, SODIUM ION | | Authors: | Yamada, K, Koutmos, M. | | Deposit date: | 2022-01-10 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | 5-Formyltetrahydrofolate promotes conformational remodeling in a methylenetetrahydrofolate reductase active site and inhibits its activity.
J.Biol.Chem., 299, 2022
|
|
7M7D
 
 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with IACS-8968 | | Descriptor: | (5S)-6,6-dimethyl-8-[(4S)-7-(trifluoromethyl)imidazo[1,5-a]pyridin-5-yl]-1,3,8-triazaspiro[4.5]decane-2,4-dione, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Leonard, P.G, Cross, J.B. | | Deposit date: | 2021-03-27 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of IACS-9779 and IACS-70465 as Potent Inhibitors Targeting Indoleamine 2,3-Dioxygenase 1 (IDO1) Apoenzyme.
J.Med.Chem., 64, 2021
|
|
5T91
 
 | | Crystal structure of B. subtilis 168 GlpQ in complex with bicine | | Descriptor: | BICINE, CALCIUM ION, Glycerophosphoryl diester phosphodiesterase, ... | | Authors: | Li, F.K.K, Strynadka, N.C.J. | | Deposit date: | 2016-09-09 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Identification of Two Phosphate Starvation-induced Wall Teichoic Acid Hydrolases Provides First Insights into the Degradative Pathway of a Key Bacterial Cell Wall Component.
J. Biol. Chem., 291, 2016
|
|
7OML
 
 | |
5C0Z
 
 | | The structure of oxidized rat cytochrome c at 1.13 angstroms resolution | | Descriptor: | Cytochrome c, somatic, HEME C, ... | | Authors: | Edwards, B.F.P, Mahapatra, G, Vaishnav, A.A, Brunzelle, J.S, Huttemann, M. | | Deposit date: | 2015-06-12 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.1236 Å) | | Cite: | Serine-47 phosphorylation of cytochromecin the mammalian brain regulates cytochromecoxidase and caspase-3 activity.
Faseb J., 2019
|
|
5TIB
 
 | | Gasdermin-B C-terminal domain containing the polymorphism residues Arg299:Ser306 fused to maltose binding protein | | Descriptor: | ACETATE ION, SODIUM ION, Sugar ABC transporter substrate-binding protein,Gasdermin-B, ... | | Authors: | Chao, K, Herzberg, O. | | Deposit date: | 2016-10-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human Gasdermin-B and disease: Sulfatide Binding, Caspase cleavage, and Structural impact of Asthma- and IBS-Associated Polymorphism
Proc.Natl.Acad.Sci.Usa, 2017
|
|
8Q6B
 
 | | The RSL-D32N - sulfonato-calix[8]arene complex, I23 form, citrate pH 4.0, obtained by cross-seeding | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, beta-D-fructopyranose, ... | | Authors: | Flood, R.J, Crowley, P.B. | | Deposit date: | 2023-08-11 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Supramolecular Synthons in Protein-Ligand Frameworks.
Cryst.Growth Des., 24, 2024
|
|
7OLH
 
 | |
7TH4
 
 | |
5T9T
 
 | | Protocadherin Gamma B2 extracellular cadherin domains 1-5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin gamma B2-alpha C, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2016-09-09 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | gamma-Protocadherin structural diversity and functional implications.
Elife, 5, 2016
|
|
5FW3
 
 | | Crystal structure of SpCas9 variant VRER bound to sgRNA and TGCG PAM target DNA | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Anders, C, Bargsten, K, Jinek, M. | | Deposit date: | 2016-02-11 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Plasticity of Pam Recognition by Engineered Variants of the RNA-Guided Endonuclease Cas9.
Mol.Cell, 61, 2016
|
|
5C26
 
 | | Crystal structure of SYK in complex with compound 1 | | Descriptor: | 3-{8-[(3,4-dimethoxyphenyl)amino]imidazo[1,2-a]pyrazin-6-yl}benzamide, GLU-VAL-PTR-GLU-SER-PRO, Tyrosine-protein kinase SYK | | Authors: | Han, S, Chang, J. | | Deposit date: | 2015-06-15 | | Release date: | 2015-10-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Imidazotriazines: Spleen Tyrosine Kinase (Syk) Inhibitors Identified by Free-Energy Perturbation (FEP).
Chemmedchem, 11, 2016
|
|
7OJR
 
 | |
8TDH
 
 | |
7TO7
 
 | |
5TKB
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 4D IN COMPLEX WITH A TETRAFLUORANLINE COMPOUND | | Descriptor: | ETHANOL, MAGNESIUM ION, N-[(2R)-2,3-dihydroxy-2-methylpropyl]-8-(methylamino)-6-[(2,3,5,6-tetrafluorophenyl)amino]imidazo[1,2-b]pyridazine-3-carboxamide, ... | | Authors: | Sack, J.S. | | Deposit date: | 2016-10-06 | | Release date: | 2016-12-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Identification of imidazo[1,2-b]pyridazine TYK2 pseudokinase ligands as potent and selective allosteric inhibitors of TYK2 signalling.
Medchemcomm, 8, 2017
|
|
8Q6A
 
 | | The RSL-D32N - sulfonato-calix[8]arene complex, I213 form, citrate pH 4.0 | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, beta-D-fructopyranose, ... | | Authors: | Flood, R.J, Crowley, P.B. | | Deposit date: | 2023-08-11 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Supramolecular Synthons in Protein-Ligand Frameworks.
Cryst.Growth Des., 24, 2024
|
|
8JZJ
 
 | | E.coli Glyceraldehyde-3-phosphate dehydrogenase structure under cryoprotect condition of ammonium sulfate | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Jang, K, Hlaing, S.H.S, Kim, H.G, Kim, N, Choe, H.W, Kim, Y.J. | | Deposit date: | 2023-07-05 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Strategy to Select an Appropriate Cryoprotectant for an X-ray Study of Escherichia coli GAPDH Crystals
Cryst.Growth Des., 23, 2023
|
|
8K9N
 
 | | Subatomic resolution structure of Pseudoazurin from Alcaligenes faecalis | | Descriptor: | COPPER (II) ION, Pseudoazurin, SULFATE ION | | Authors: | Fukuda, Y, Lintuluoto, M, Kurihara, K, Hasegawa, K, Inoue, T, Tamada, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Overlooked Hydrogen Bond in a Blue Copper Protein Uncovered by Neutron and Sub- angstrom ngstrom Resolution X-ray Crystallography.
Biochemistry, 63, 2024
|
|
8TS0
 
 | |
5FVN
 
 | | X-ray crystal structure of Enterobacter cloacae OmpE36 porin. | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 3-HYDROXY-TETRADECANOIC ACID, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, ... | | Authors: | Arunmanee, W, Pathania, M, Soloyova, A, Brun, A, Ridley, H, Basle, A, van den Berg, B, Lakey, J.H. | | Deposit date: | 2016-02-09 | | Release date: | 2016-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Gram-negative trimeric porins have specific LPS binding sites that are essential for porin biogenesis.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
