2XJ0
 
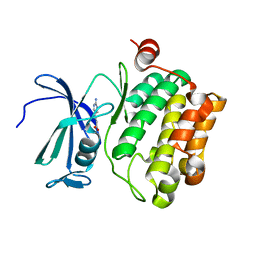 | | Protein kinase Pim-1 in complex with fragment-4 from crystallographic fragment screen | | Descriptor: | (E)-3-(2-AMINO-PYRIDINE-5YL)-ACRYLIC ACID, PROTO-ONCOGENE SERINE/THREONINE PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
6NAY
 
 | |
6ND4
 
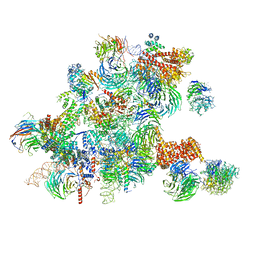 | | Conformational switches control early maturation of the eukaryotic small ribosomal subunit | | Descriptor: | 18S rRNA 5' domain start, 5'ETS rRNA, Bud21, ... | | Authors: | Hunziker, M, Barandun, J, Klinge, S. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Conformational switches control early maturation of the eukaryotic small ribosomal subunit.
Elife, 8, 2019
|
|
6NAW
 
 | |
8F39
 
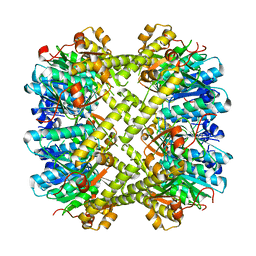
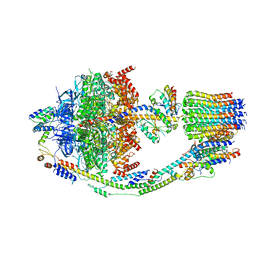 | | Yeast ATP synthase in conformation-2, at pH 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase protein 8, ATP synthase subunit 4, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-11-09 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8F29
 
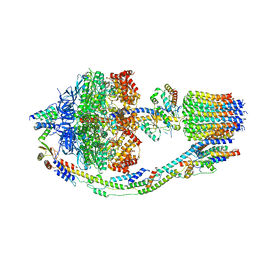 | | Yeast ATP synthase in conformation-1 at pH 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase protein 8, ATP synthase subunit 4, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-11-07 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3RZS
 
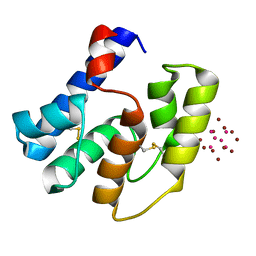 | | Apis mellifera OBP14 in complex with Ta6Br14 | | Descriptor: | HEXATANTALUM DODECABROMIDE, OBP14 | | Authors: | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-05-12 | | Release date: | 2011-11-30 | | Last modified: | 2012-01-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
3S0A
 
 | | Apis mellifera OBP14, native apo-protein | | Descriptor: | OBP14 | | Authors: | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
6NF8
 
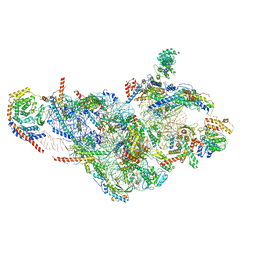 | | Structure of human mitochondrial translation initiation factor 3 bound to the small ribosomal subunit -Class I | | Descriptor: | 28S ribosomal RNA, mitochondria, 28S ribosomal protein S10, ... | | Authors: | Sharma, M, Koripella, R, Agrawal, R. | | Deposit date: | 2018-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structure of Human Mitochondrial Translation Initiation Factor 3 Bound to the Small Ribosomal Subunit.
iScience, 12, 2019
|
|
6NER
 
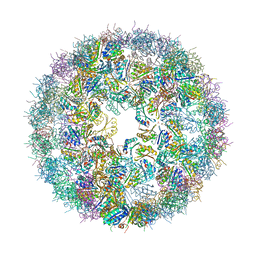 | | Synthetic Haliangium ochraceum BMC shell | | Descriptor: | BMC-H tandem fusion protein, SULFATE ION | | Authors: | Sutter, M, McGuire, S, Aussignargues, C, Kerfeld, C.A. | | Deposit date: | 2018-12-18 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural Characterization of a Synthetic Tandem-Domain Bacterial Microcompartment Shell Protein Capable of Forming Icosahedral Shell Assemblies.
ACS Synth Biol, 8, 2019
|
|
4OTO
 
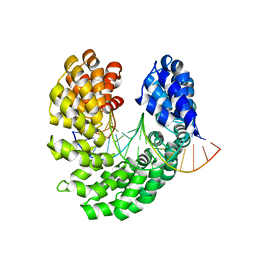 | | Crystal structure of the S505W mutant of TAL effector dHax3 | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*TP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*AP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3 | | Authors: | Deng, D, Wu, J.P, Yan, C.Y, Pan, X.J, Yan, N. | | Deposit date: | 2014-02-14 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.588 Å) | | Cite: | Revisiting the TALE repeat
Protein Cell, 5, 2014
|
|
4OSZ
 
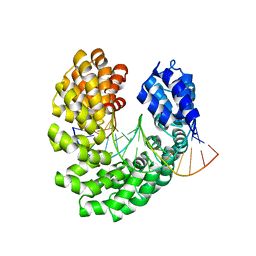 | | Crystal structure of the S505P mutant of TAL effector dHax3 | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*TP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*AP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3 | | Authors: | Deng, D, Wu, J.P, Yan, C.Y, Pan, X.J, Yan, N. | | Deposit date: | 2014-02-13 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.614 Å) | | Cite: | Revisiting the TALE repeat
Protein Cell, 5, 2014
|
|
6NEQ
 
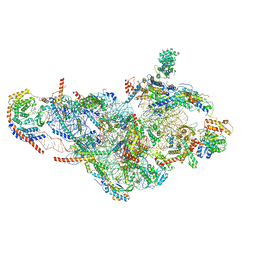 | | Structure of human mitochondrial translation initiation factor 3 bound to the small ribosomal subunit-Class-II | | Descriptor: | 28S ribosomal RNA, mitochondrial, 28S ribosomal protein S10, ... | | Authors: | Sharma, M, Koripella, R, Agrawal, R. | | Deposit date: | 2018-12-18 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structure of Human Mitochondrial Translation Initiation Factor 3 Bound to the Small Ribosomal Subunit.
iScience, 12, 2019
|
|
6NHG
 
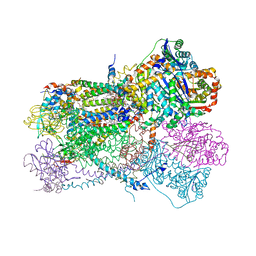 | | Rhodobacter sphaeroides Mitochondrial respiratory chain complex | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl octadecanoate, 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Xia, D, Zhou, F, Esser, L. | | Deposit date: | 2018-12-21 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of bacterial cytochromebc1in complex with azoxystrobin reveals a conformational switch of the Rieske iron-sulfur protein subunit.
J.Biol.Chem., 294, 2019
|
|
3T5J
 
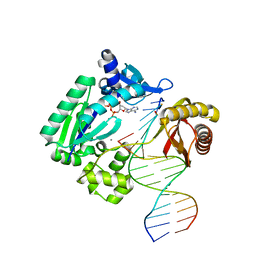 | | Ternary complex of HNE Adduct modified DNA (5'-TXG-3' vs 13-mer) with Dpo4 and incoming dDTP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*CP*AP*TP*(HN1)P*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Banerjee, S, Christov, P.P, Kozekova, A, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2011-07-27 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Replication bypass of the trans-4-Hydroxynonenal-derived (6S,8R,11S)-1,N(2)-deoxyguanosine DNA adduct by the sulfolobus solfataricus DNA polymerase IV.
Chem.Res.Toxicol., 25, 2012
|
|
6DR4
 
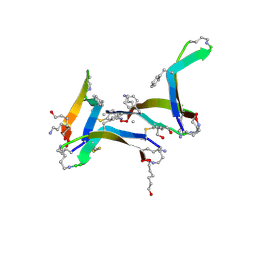 | |
4P2R
 
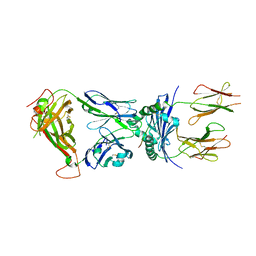 | | Crystal structure of the 5cc7 TCR in complex with 5c1/I-Ek | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5c1 peptide, 5cc7 T-cell receptor alpha chain, ... | | Authors: | Birnbaum, M.E, Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-03-04 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.295 Å) | | Cite: | Deconstructing the Peptide-MHC Specificity of T Cell Recognition.
Cell, 157, 2014
|
|
4P70
 
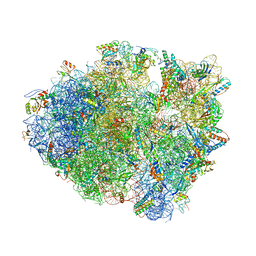 | | Crystal Structure of Unmodified tRNA Proline (CGG) Bound to Codon CCG on the Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2014-03-25 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.68 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2XRO
 
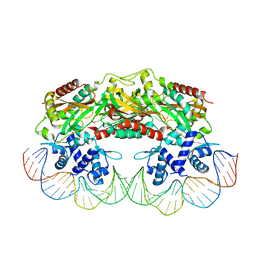 | | Crystal structure of TtgV in complex with its DNA operator | | Descriptor: | HTH-TYPE TRANSCRIPTIONAL REGULATOR TTGV, OSMIUM ION, TTGV OPERATOR DNA | | Authors: | Lu, D, Fillet, S, Meng, C, Alguel, Y, Kloppsteck, P, Bergeron, J, Krell, T, Gallegos, M.-T, Ramos, J, Zhang, X. | | Deposit date: | 2010-09-17 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of Ttgv in Complex with its DNA Operator Reveals a General Model for Cooperative DNA Binding of Tetrameric Gene Regulators.
Genes Dev., 24, 2010
|
|
8FR8
 
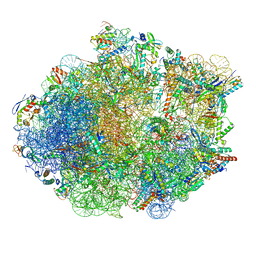 | | Structure of Mycobacterium smegmatis Rsh bound to a 70S translation initiation complex | | Descriptor: | 16S rRNA (1511-MER), 23S rRNA (3119-MER), 30S ribosomal protein S10, ... | | Authors: | Majumdar, S, Sharma, M.R, Manjari, S.R, Banavali, N.K, Agrawal, R.K. | | Deposit date: | 2023-01-06 | | Release date: | 2023-05-17 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Starvation sensing by mycobacterial RelA/SpoT homologue through constitutive surveillance of translation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2XIX
 
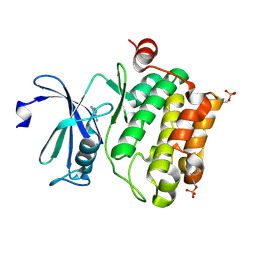 | | Protein kinase Pim-1 in complex with fragment-1 from crystallographic fragment screen | | Descriptor: | 3,5-DIAMINO-1H-[1,2,4]TRIAZOLE, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XRZ
 
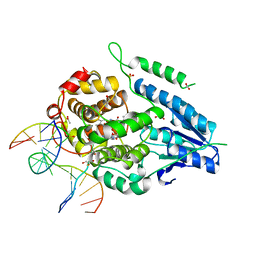 | | X-ray structure of archaeal class II CPD photolyase from Methanosarcina mazei in complex with intact CPD-lesion | | Descriptor: | ACETATE ION, COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, ... | | Authors: | Kiontke, S, Geisselbrecht, Y, Pokorny, R, Carell, T, Batschauer, A, Essen, L.O. | | Deposit date: | 2010-09-24 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of an Archaeal Class II DNA Photolyase and its Complex with Uv-Damaged Duplex DNA.
Embo J., 30, 2011
|
|
8FLF
 
 | |
2XIY
 
 | | Protein kinase Pim-1 in complex with fragment-2 from crystallographic fragment screen | | Descriptor: | 2-HYDROXYMETHYL-BENZOIMIDAZOLE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
6NAQ
 
 | | Crystal structure of Neisseria meningitidis ClpP protease in Apo form | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, POTASSIUM ION | | Authors: | Houry, W.A, Mabanglo, M.F, Pai, E.F, Eger, B.T, Bryson, S. | | Deposit date: | 2018-12-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.022 Å) | | Cite: | ClpP protease activation results from the reorganization of the electrostatic interaction networks at the entrance pores.
Commun Biol, 2, 2019
|
|
