2TDM
 
 | | STRUCTURE OF THYMIDYLATE SYNTHASE | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1997-03-28 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Refined structures of substrate-bound and phosphate-bound thymidylate synthase from Lactobacillus casei.
J.Mol.Biol., 232, 1993
|
|
6HD7
 
 | | Cryo-EM structure of the ribosome-NatA complex | | Descriptor: | 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, 5.8S rRNA, 5S rRNA, ... | | Authors: | Knorr, A.G, Becker, T, Beckmann, R. | | Deposit date: | 2018-08-17 | | Release date: | 2018-12-19 | | Last modified: | 2019-01-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Ribosome-NatA architecture reveals that rRNA expansion segments coordinate N-terminal acetylation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
6JR1
 
 | | Crystal structure of the human nucleosome phased with 16 selenium atoms | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Saotome, M, Horikoshi, N, Urano, K, Kujirai, T, Yuzurihara, H, Kurumizaka, H, Kagawa, W. | | Deposit date: | 2019-04-02 | | Release date: | 2019-10-02 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure determination of the nucleosome core particle by selenium SAD phasing.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6I84
 
 | | Structure of transcribing RNA polymerase II-nucleosome complex | | Descriptor: | DNA (158-MER), DNA (170-MER), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Farnung, L, Vos, S.M, Cramer, P. | | Deposit date: | 2018-11-19 | | Release date: | 2019-01-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of transcribing RNA polymerase II-nucleosome complex.
Nat Commun, 9, 2018
|
|
6JR0
 
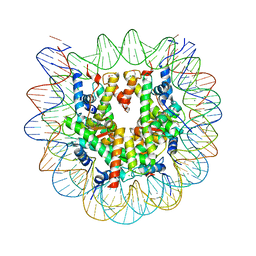 | | Crystal structure of the human nucleosome phased with 12 selenium atoms | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Saotome, M, Horikoshi, N, Urano, K, Kujirai, T, Yuzurihara, H, Kurumizaka, H, Kagawa, W. | | Deposit date: | 2019-04-02 | | Release date: | 2019-10-02 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure determination of the nucleosome core particle by selenium SAD phasing.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7WWG
 
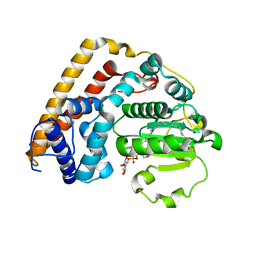 | | Crystal structure of Saccharomyces cerevisiae Sfh2 complexed with phosphatidylinositol in an open conformation | | Descriptor: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Phosphatidylinositol transfer protein CSR1 | | Authors: | Chen, L, Tan, L, Im, Y.J. | | Deposit date: | 2022-02-12 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis of ligand recognition and transport by Sfh2, a yeast phosphatidylinositol transfer protein of the Sec14 superfamily.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7X22
 
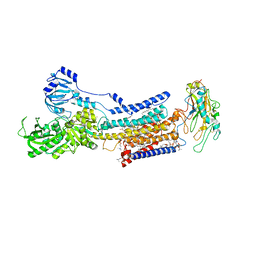 | | Cryo-EM structure of non gastric H,K-ATPase alpha2 K794S in (2K+)E2-AlF state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Nakanishi, H, Abe, K. | | Deposit date: | 2022-02-25 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and function of H + /K + pump mutants reveal Na + /K + pump mechanisms.
Nat Commun, 13, 2022
|
|
7WVT
 
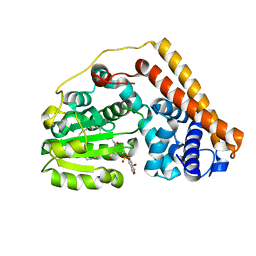 | | Crystal structure of Saccharomyces cerevisiae Sfh2 complexed with phosphatidylinositol | | Descriptor: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Phosphatidylinositol transfer protein CSR1 | | Authors: | Chen, L, Tan, L, Im, Y.J. | | Deposit date: | 2022-02-11 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of ligand recognition and transport by Sfh2, a yeast phosphatidylinositol transfer protein of the Sec14 superfamily.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6Z00
 
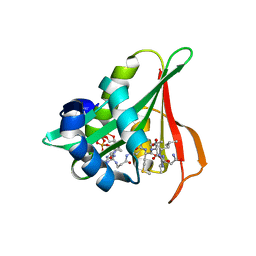 | | Arabidopsis thaliana Naa50 in complex with bisubstrate analogue CoA-Ac-MVNAL | | Descriptor: | Acyl-CoA N-acyltransferases (NAT) superfamily protein, CARBOXYMETHYL COENZYME *A, MET-VAL-ASN-ALA-LEU | | Authors: | Weidenhausen, J, Kopp, J, Lapouge, K, Sinning, I. | | Deposit date: | 2020-05-07 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural and functional characterization of the N-terminal acetyltransferase Naa50.
Structure, 29, 2021
|
|
6YZZ
 
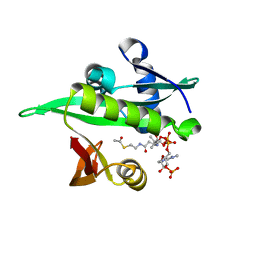 | | Arabidopsis thaliana Naa50 in complex with AcCoA | | Descriptor: | ACETYL COENZYME *A, N-alpha-acetyltransferase 50 | | Authors: | Weidenhausen, J, Kopp, J, Lapouge, K, Sinning, I. | | Deposit date: | 2020-05-07 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural and functional characterization of the N-terminal acetyltransferase Naa50.
Structure, 29, 2021
|
|
6ZFB
 
 | | Structure of the B. subtilis RNA POLYMERASE in complex with HelD (dimer) | | Descriptor: | DNA helicase, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Pei, H.-P, Hilal, T, Huang, Y.-H, Said, N, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The delta subunit and NTPase HelD institute a two-pronged mechanism for RNA polymerase recycling.
Nat Commun, 11, 2020
|
|
5DKE
 
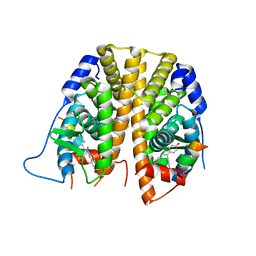 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a 3-naphthyl-substituted, methyl, cis-diaryl-ethylene compound 4,4'-[2-(naphthalen-2-yl)prop-1-ene-1,1-diyl]diphenol | | Descriptor: | 4,4'-[2-(naphthalen-2-yl)prop-1-ene-1,1-diyl]diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-03 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DL4
 
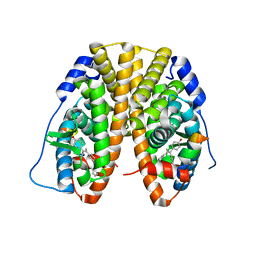 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a phenylamino-substituted, methyl, triaryl-ethylene derivative 4,4'-{2-[3-(phenylamino)phenyl]prop-1-ene-1,1-diyl}diphenol | | Descriptor: | 4,4'-{2-[3-(phenylamino)phenyl]prop-1-ene-1,1-diyl}diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
7X20
 
 | |
7X24
 
 | | Cryo-EM structure of non gastric H,K-ATPase alpha2 SPWC mutant in (2K+)E2-AlF state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Abe, K, Nakanishi, H, Young, V, Artigas, P. | | Deposit date: | 2022-02-25 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and function of H + /K + pump mutants reveal Na + /K + pump mechanisms.
Nat Commun, 13, 2022
|
|
5DTV
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a dimethyl-substituted, 3,4-diarylthiophene dioxide core ligand | | Descriptor: | 3,4-bis(4-hydroxy-2-methylphenyl)-1H-1lambda~6~-thiophene-1,1-dione, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-18 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
7X21
 
 | | Cryo-EM structure of non gastric H,K-ATPase alpha2 K794A in (K+)E2-AlF state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Nakanishi, H, Abe, K. | | Deposit date: | 2022-02-25 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and function of H + /K + pump mutants reveal Na + /K + pump mechanisms.
Nat Commun, 13, 2022
|
|
6GX7
 
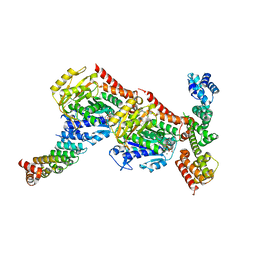 | | Tubulin-CopN-alphaRep complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gigant, B, Campanacci, V. | | Deposit date: | 2018-06-26 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Insight into microtubule nucleation from tubulin-capping proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7X23
 
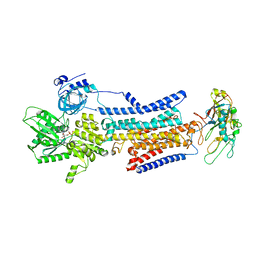 | | Cryo-EM structure of non gastric H,K-ATPase alpha2 SPWC mutant in 3Na+E1-AMPPCPF state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Potassium-transporting ATPase alpha chain 2, ... | | Authors: | Abe, K, Nakanishi, H, Young, V, Artigas, P. | | Deposit date: | 2022-02-25 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and function of H + /K + pump mutants reveal Na + /K + pump mechanisms.
Nat Commun, 13, 2022
|
|
8J07
 
 | |
5DRM
 
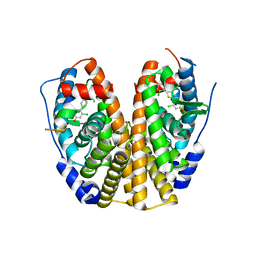 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a dichloro-substituted, 2,5-diarylthiophene-core ligand 4,4'-thiene-2,5-diylbis(3-chlorophenol) | | Descriptor: | 4,4'-thiene-2,5-diylbis(3-chlorophenol), Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-16 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.241 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DKG
 
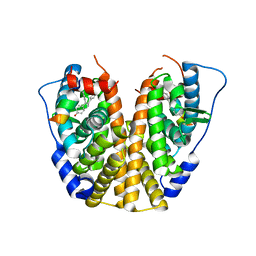 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a t-butyl-substituted, methyl, triaryl-ethylene derivative 4,4'-[2-(4-tert-butylphenyl)prop-1-ene-1,1-diyl]diphenol | | Descriptor: | 4,4'-[2-(4-tert-butylphenyl)prop-1-ene-1,1-diyl]diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-03 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DVS
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with a 2-Methyl-substituted Triaryl-imine 4,4'-[(2-methylphenyl)carbonimidoyl]diphenol | | Descriptor: | 4,4'-[(2-methylphenyl)carbonimidoyl]diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Wright, N.J, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-21 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
6GVN
 
 | | Tubulin:TM-3 DARPin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gigant, B, Cantos Fernandes, S, Campanacci, V. | | Deposit date: | 2018-06-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Insight into microtubule nucleation from tubulin-capping proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5DMF
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a 4-fluorophenylamino-substituted, methyl triaryl-ethylene derivative 4,4'-(2-{3-[(4-fluorophenyl)amino]phenyl}prop-1-ene-1,1-diyl)diphenol | | Descriptor: | 4,4'-(2-{3-[(4-fluorophenyl)amino]phenyl}prop-1-ene-1,1-diyl)diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
