5WP6
 
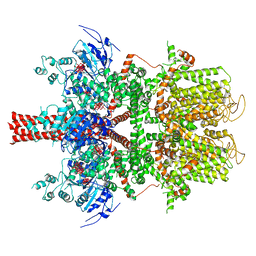 | | Cryo-EM structure of a human TRPM4 channel in complex with calcium and decavanadate | | Descriptor: | DECAVANADATE, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Winkler, P.A, Huang, Y, Sun, W, Du, J, Lu, W. | | Deposit date: | 2017-08-03 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Electron cryo-microscopy structure of a human TRPM4 channel.
Nature, 552, 2017
|
|
6OP4
 
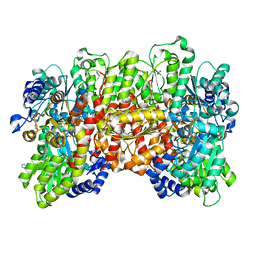 | | Selenium-incorporated, carbon monoxide-inhibited, reactivated FeMo-cofactor of nitrogenase from Azotobacter vinelandii | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Arias, R.J, Rees, D.C. | | Deposit date: | 2019-04-24 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Localized Electronic Structure of Nitrogenase FeMoco Revealed by Selenium K-Edge High Resolution X-ray Absorption Spectroscopy.
J.Am.Chem.Soc., 141, 2019
|
|
5E4X
 
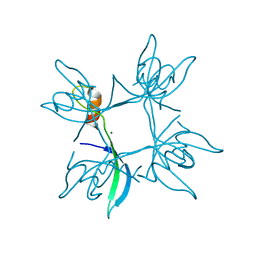 | | Crystal structure of cpSRP43 chromodomain 3 | | Descriptor: | MAGNESIUM ION, Signal recognition particle 43 kDa protein, chloroplastic | | Authors: | Horn, A, Ahmed, Y.L, Wild, K, Sinning, I. | | Deposit date: | 2015-10-07 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for cpSRP43 chromodomain selectivity and dynamics in Alb3 insertase interaction.
Nat Commun, 6, 2015
|
|
4ON9
 
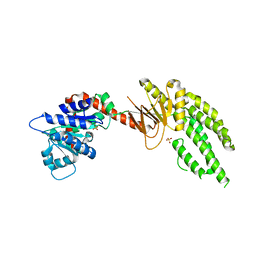 | | DECH box helicase domain | | Descriptor: | CHLORIDE ION, Probable ATP-dependent RNA helicase DDX58, SULFATE ION | | Authors: | Deimling, T, Witte, G, Hopfner, K.P. | | Deposit date: | 2014-01-28 | | Release date: | 2014-07-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal and solution structure of the human RIG-I SF2 domain
Acta Crystallogr.,Sect.F, 70, 2014
|
|
5X2J
 
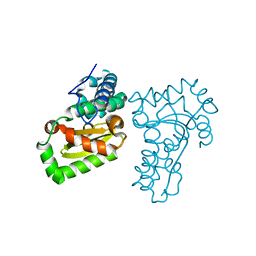 | | Crystal structure of a recombinant hybrid manganese superoxide dismutase from Staphylococcus equorum and Staphylococcus saprophyticus | | Descriptor: | MANGANESE (II) ION, manganese superoxide dismutase | | Authors: | Retnoningrum, D.S, Yoshida, H, Arumsari, S, Kamitori, S, Ismaya, W.T. | | Deposit date: | 2017-01-31 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The first crystal structure of manganese superoxide dismutase from the genus Staphylococcus
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3H1P
 
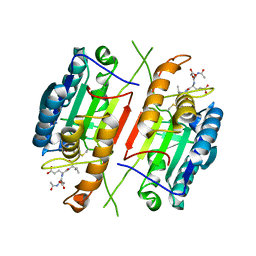 | |
3ADI
 
 | |
2B4M
 
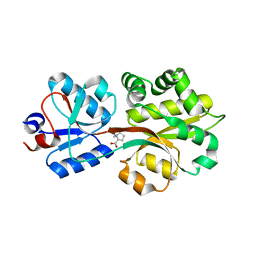 | | Crystal structure of the binding protein OpuAC in complex with proline betaine | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, Glycine betaine-binding protein | | Authors: | Horn, C, Sohn-Boesser, L, Breed, J, Welte, W, Schmitt, L, Bremer, E. | | Deposit date: | 2005-09-26 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Determinants for Substrate Specificity of the Ligand-binding Protein OpuAC from Bacillus subtilis for the Compatible Solutes Glycine Betaine and Proline Betaine.
J.Mol.Biol., 357, 2006
|
|
2XOD
 
 | | Crystal structure of flavoprotein NrdI from Bacillus anthracis in the oxidised form | | Descriptor: | CACODYLATE ION, FLAVIN MONONUCLEOTIDE, NRDI PROTEIN, ... | | Authors: | Johansson, R, Sprenger, J, Torrents, E, Sahlin, M, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2010-08-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | High Resolution Crystal Structures of Nrdi in the Oxidised and Reduced States: An Unusual Flavodoxin
FEBS J., 277, 2010
|
|
4FHC
 
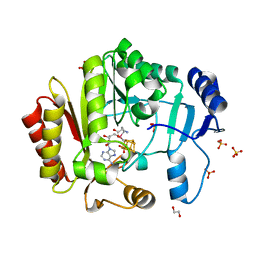 | | Spore photoproduct lyase | | Descriptor: | 1,2-ETHANEDIOL, IRON/SULFUR CLUSTER, SULFATE ION, ... | | Authors: | Benjdia, A, Heil, K, Barends, T.R.M, Carell, T, Schlichting, I. | | Deposit date: | 2012-06-06 | | Release date: | 2012-07-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into recognition and repair of UV-DNA damage by Spore Photoproduct Lyase, a radical SAM enzyme.
Nucleic Acids Res., 40, 2012
|
|
3ADL
 
 | |
4O9G
 
 | | Crystal structure of the H51N mutant of the 3,4-ketoisomerase QdtA from Thermoanaerobacterium thermosaccharolyticum in complex with TDP-4-keto-6-deoxyglucose | | Descriptor: | (2S)-1-[3-[(2S)-2-oxidanylpropoxy]-2-[[(2S)-2-oxidanylpropoxy]methyl]-2-[[(2R)-2-oxidanylpropoxy]methyl]propoxy]propan-2-ol, 1,2-ETHANEDIOL, QdtA, ... | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2014-01-02 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The molecular architecture of QdtA, a sugar 3,4-ketoisomerase from Thermoanaerobacterium thermosaccharolyticum.
Protein Sci., 23, 2014
|
|
5Z69
 
 | |
5AKO
 
 | |
4OX3
 
 | | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition | | Descriptor: | PHOSPHATE ION, Putative carboxypeptidase YodJ, ZINC ION | | Authors: | Hoyland, C.N, Aldridge, C, Cleverley, R.M, Sidiq, K, Duchene, M.C, Daniel, R.A, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-02-04 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition.
Structure, 22, 2014
|
|
3HU3
 
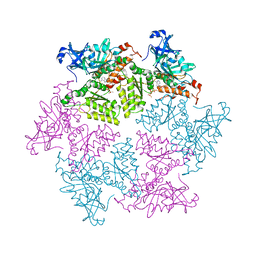 | | Structure of p97 N-D1 R155H mutant in complex with ATPgS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Tang, W.-K. | | Deposit date: | 2009-06-12 | | Release date: | 2010-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants.
Embo J., 29, 2010
|
|
5HKE
 
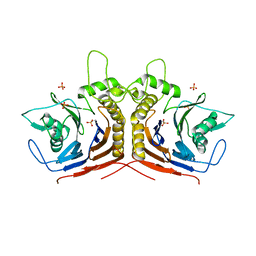 | | bile salt hydrolase from Lactobacillus salivarius | | Descriptor: | Bile salt hydrolase, PHOSPHATE ION | | Authors: | Hu, X.-J. | | Deposit date: | 2016-01-14 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of bile salt hydrolase from Lactobacillus salivarius.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
1GQN
 
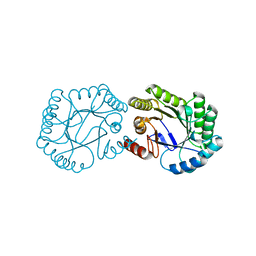 | | Native 3-dehydroquinase from Salmonella typhi | | Descriptor: | 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Lee, W.-H, Perles, L.A, Nagem, R.A.P, Polikarpov, I, Sawyer, L. | | Deposit date: | 2001-11-26 | | Release date: | 2003-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Comparison of Different Crystal Forms of 3-Dehydroquinase from Salmonella Typhi and its Implications for Enzyme Activity
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3GZL
 
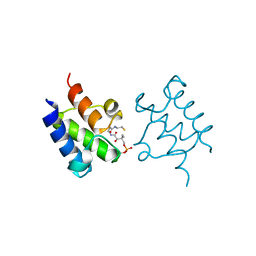 | | Crystal Structure of holo PfACP Disulfide-Linked Dimer | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein | | Authors: | Gallagher, J.R, Prigge, S.T. | | Deposit date: | 2009-04-07 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Plasmodium falciparum acyl carrier protein crystal structures in disulfide-linked and reduced states and their prevalence during blood stage growth.
Proteins, 78, 2010
|
|
1GY5
 
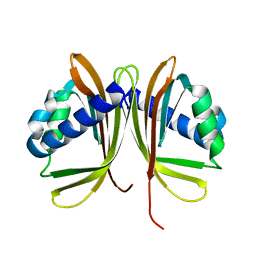 | |
2H1F
 
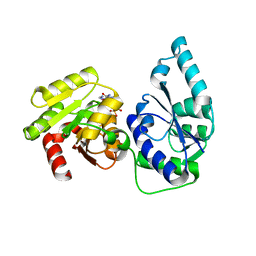 | | E. coli heptosyltransferase WaaC with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lipopolysaccharide heptosyltransferase-1 | | Authors: | Grizot, S, Salem, M, Vongsouthi, V, Durand, L, Moreau, F, Dohi, H, Vincent, S, Escaich, S, Ducruix, A. | | Deposit date: | 2006-05-16 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Escherichia coli heptosyltransferase WaaC: binary complexes with ADP and ADP-2-deoxy-2-fluoro heptose.
J.Mol.Biol., 363, 2006
|
|
2EPL
 
 | | N-acetyl-B-D-glucosaminidase (GCNA) from Streptococcus gordonii | | Descriptor: | GLYCEROL, N-acetyl-beta-D-glucosaminidase, SULFATE ION | | Authors: | Langley, D.B, Harty, D.W.S, Guss, J.M, Collyer, C.A. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of N-acetyl-beta-D-glucosaminidase (GcnA) from the Endocarditis Pathogen Streptococcus gordonii and its Complex with the Mechanism-based Inhibitor NAG-thiazoline
J.Mol.Biol., 377, 2008
|
|
3H16
 
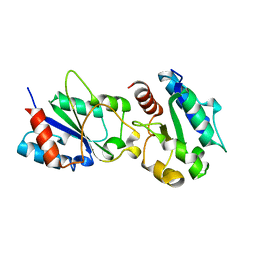 | | Crystal structure of a bacteria TIR domain, PdTIR from Paracoccus denitrificans | | Descriptor: | SULFATE ION, TIR protein | | Authors: | Chan, S.L, Low, L.Y, Santelli, E, Pascual, J. | | Deposit date: | 2009-04-11 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Mimicry in Innate Immunity: CRYSTAL STRUCTURE OF A BACTERIAL TIR DOMAIN.
J.Biol.Chem., 284, 2009
|
|
5A4W
 
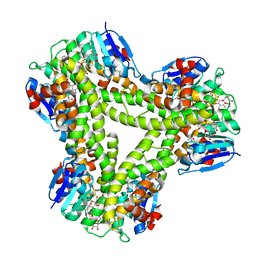 | | AtGSTF2 from Arabidopsis thaliana in complex with quercetrin | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4-oxo-4H-chromen-3-yl 6-deoxy-alpha-L-mannopyranoside, ACETATE ION, GLUTATHIONE S-TRANSFERASE F2 | | Authors: | Ahmad, L, Rylott, E, Bruce, N.C, Edwards, R, Grogan, G. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural evidence for Arabidopsis glutathione transferase AtGSTF2 functioning as a transporter of small organic ligands.
FEBS Open Bio, 7, 2017
|
|
1H5B
 
 | | T cell receptor Valpha11 (AV11S5) domain | | Descriptor: | CHLORIDE ION, GLYCEROL, MURINE T CELL RECEPTOR (TCR) VALPHA DOMAIN | | Authors: | Machius, M, Cianga, P, Deisenhofer, J, Sally Ward, E. | | Deposit date: | 2001-05-21 | | Release date: | 2001-06-21 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of a T Cell Receptor Valpha11 (Av11S5) Domain: New Canonical Forms for the First and Second Complementarity Determining Regions
J.Mol.Biol., 310, 2001
|
|
