2EU7
 
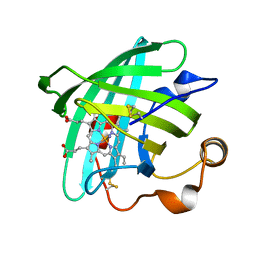 | |
2A69
 
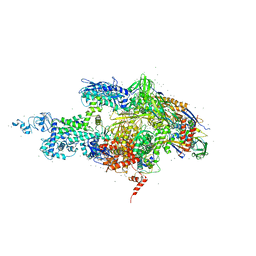 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with antibiotic rifapentin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
2EKT
 
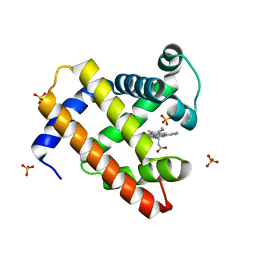 | | Crystal structure of myoglobin reconstituted with 6-methyl-6-depropionatehemin | | Descriptor: | 6-METHY-6-DEPROPIONATEHEMIN, Myoglobin, SULFATE ION | | Authors: | Harada, K, Makino, M, Sugimoto, H, Hirota, S, Matsuo, T, Shiro, Y, Hisaeda, Y, Hayashi, T. | | Deposit date: | 2007-03-25 | | Release date: | 2007-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure and ligand binding properties of myoglobins reconstituted with monodepropionated heme: functional role of each heme propionate side chain
Biochemistry, 46, 2007
|
|
2ERP
 
 | | Crystal structure of vascular apoptosis-inducing protein-1(inhibitor-bound form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(N-HYDROXYCARBOXAMIDO)-2-ISOBUTYLPROPANOYL-TRP-METHYLAMIDE, CALCIUM ION, ... | | Authors: | Takeda, S, Igarashi, T, Araki, S. | | Deposit date: | 2005-10-25 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structures of VAP1 reveal ADAMs' MDC domain architecture and its unique C-shaped scaffold
Embo J., 25, 2006
|
|
1DD7
 
 | |
1SRE
 
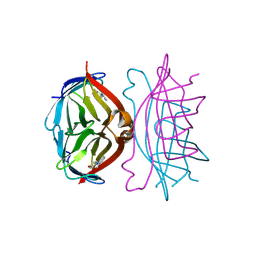 | |
2BZB
 
 | | NMR Solution Structure of a protein aspartic acid phosphate phosphatase from Bacillus Anthracis | | Descriptor: | CONSERVED DOMAIN PROTEIN | | Authors: | Grenha, R, Rzechorzek, N.J, Brannigan, J.A, Ab, E, Folkers, G.E, De Jong, R.N, Diercks, T, Wilkinson, A.J, Kaptein, R, Wilson, K.S. | | Deposit date: | 2005-08-14 | | Release date: | 2006-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of Spo0E-like protein-aspartic acid phosphatases that regulate sporulation in bacilli.
J. Biol. Chem., 281, 2006
|
|
2BW2
 
 | | BofC from Bacillus subtilis | | Descriptor: | BYPASS OF FORESPORE C | | Authors: | Patterson, H.M, Brannigan, J.A, Cutting, S.M, Wilson, K.S, Wilkinson, A.J, Ab, E, Diercks, T, Folkers, G.E, de Jong, R.N, Truffault, V, Kaptein, R. | | Deposit date: | 2005-07-08 | | Release date: | 2005-09-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of bypass of forespore C, an intercompartmental signaling factor during sporulation in Bacillus.
J. Biol. Chem., 280, 2005
|
|
2BJB
 
 | | Mycobacterium Tuberculosis Epsp Synthase In Unliganded State | | Descriptor: | 3-PHOSPHOSHIKIMATE 1-CARBOXYVINYLTRANSFERASE, ACETATE ION, SODIUM ION | | Authors: | Bourenkov, G.P, Kachalova, G.S, Strizhov, N, Bruning, M, Vagin, A, Bartunik, H.D. | | Deposit date: | 2005-02-01 | | Release date: | 2006-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mycobacterium Tuberculosis Epsp Synthase in Unliganded State
To be Published
|
|
2BFE
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-12-06 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2BFB
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-12-06 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2BFD
 
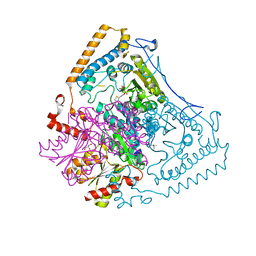 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-12-06 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2C40
 
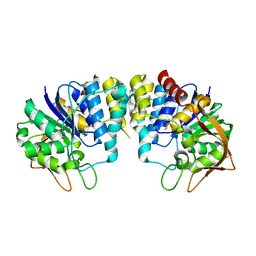 | | CRYSTAL STRUCTURE OF INOSINE-URIDINE PREFERRING NUCLEOSIDE HYDROLASE FROM BACILLUS ANTHRACIS AT 2.2A RESOLUTION | | Descriptor: | CALCIUM ION, INOSINE-URIDINE PREFERRING NUCLEOSIDE HYDROLASE FAMILY PROTEIN, alpha-D-ribofuranose | | Authors: | Moroz, O.V, Blagova, E.V, Fogg, M.J, Levdikov, V.M, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2005-10-13 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Inosine-Uridine Preferring Nucleoside Hydrolase from Bacillus Anthracis at 2.2A Resolution
To be Published
|
|
2BEU
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, C2-1-HYDROXY-3-METHYL-PROPYL-THIAMIN DIPHOSPHATE, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-11-30 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2C29
 
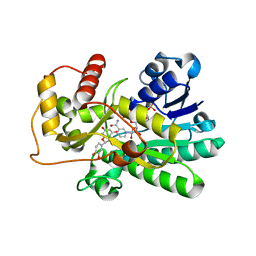 | | Structure of dihydroflavonol reductase from Vitis vinifera at 1.8 A. | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, DIHYDROFLAVONOL 4-REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petit, P, Granier, T, D'Estaintot, B.L, Hamdi, S, Gallois, B. | | Deposit date: | 2005-09-27 | | Release date: | 2006-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structure of Grape Dihydroflavonol 4-Reductase, a Key Enzyme in Flavonoid Biosynthesis.
J.Mol.Biol., 368, 2007
|
|
2BEW
 
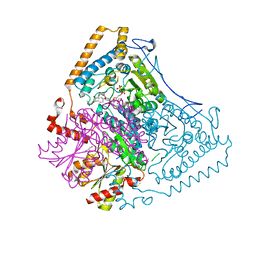 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, C2-1-HYDROXYPHENYL-THIAMIN DIPHOSPHATE, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-11-30 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2BFC
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-12-06 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
7K15
 
 | | Crystal structure of the Human Leukotriene B4 Receptor 1 in Complex with Selective Antagonist MK-D-046 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, FLAVIN MONONUCLEOTIDE, HEXAETHYLENE GLYCOL, ... | | Authors: | Michaelian, N, Han, G.W, Cherezov, V. | | Deposit date: | 2020-09-07 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural insights on ligand recognition at the human leukotriene B4 receptor 1.
Nat Commun, 12, 2021
|
|
1N72
 
 | | Structure and Ligand of a Histone Acetyltransferase Bromodomain | | Descriptor: | HISTONE ACETYLTRANSFERASE | | Authors: | Dhalluin, C, Carlson, J.E, Zeng, L, He, C, Aggarwal, A.K, Zhou, M.-M. | | Deposit date: | 2002-11-12 | | Release date: | 2002-12-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and Ligand of a Histone Acetyltransferase Bromodomain
Nature, 399, 1999
|
|
1E5C
 
 | | Internal xylan binding domain from C. fimi Xyn10A, R262G mutant | | Descriptor: | XYLANASE D | | Authors: | Simpson, P.J, Hefang, X, Bolam, D.N, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2000-07-24 | | Release date: | 2001-05-25 | | Last modified: | 2018-10-24 | | Method: | SOLUTION NMR | | Cite: | The Structural Basis for the Ligand Specificity of Family 2 Carbohydrate Binding Nodules
J.Biol.Chem., 275, 2000
|
|
1EYV
 
 | | THE CRYSTAL STRUCTURE OF NUSB FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | N-UTILIZING SUBSTANCE PROTEIN B HOMOLOG, PHOSPHATE ION | | Authors: | Gopal, B, Haire, L.F, Cox, R.A, Colston, M.J, Major, S, Brannigan, J.A, Smerdon, S.J, Dodson, G.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-05-09 | | Release date: | 2000-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of NusB from Mycobacterium tuberculosis.
Nat.Struct.Biol., 7, 2000
|
|
2C0D
 
 | | Structure of the mitochondrial 2-cys peroxiredoxin from Plasmodium falciparum | | Descriptor: | THIOREDOXIN PEROXIDASE 2 | | Authors: | Boucher, I.W, Brannigan, J.A, Wilkinson, A.J, Brzozowski, A.M, Muller, S. | | Deposit date: | 2005-09-01 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and Biochemical Characterisation of a Mitochondrial Peroxiredoxin from Plasmodium Falciparum
Mol.Microbiol., 61, 2006
|
|
2C20
 
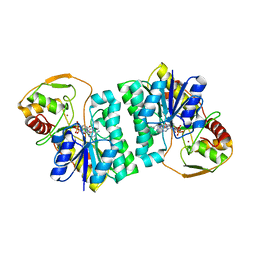 | | CRYSTAL STRUCTURE OF UDP-GLUCOSE 4-EPIMERASE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-GLUCOSE 4-EPIMERASE, ZINC ION | | Authors: | Lebedev, A.A, Moroz, O.V, Blagova, E.V, Levdikov, V.M, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2005-09-22 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Udp-Glucose 4-Epimerase from Bacillus Anthracis at 2.7A Resolution
To be Published
|
|
2C8I
 
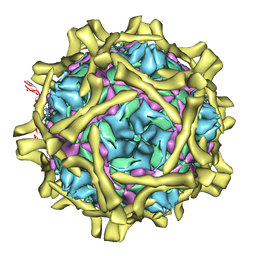 | | Complex Of Echovirus Type 12 With Domains 1, 2, 3 and 4 Of Its Receptor Decay Accelerating Factor (Cd55) By Cryo Electron Microscopy At 16 A | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR, ECHOVIRUS 11 COAT PROTEIN VP1, ECHOVIRUS 11 COAT PROTEIN VP2, ... | | Authors: | Pettigrew, D.M, Williams, D.T, Kerrigan, D, Evans, D.J, Lea, S.M, Bhella, D. | | Deposit date: | 2005-12-05 | | Release date: | 2006-01-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Structural and Functional Insights Into the Interaction of Echoviruses and Decay-Accelerating Factor.
J.Biol.Chem., 281, 2006
|
|
1E6O
 
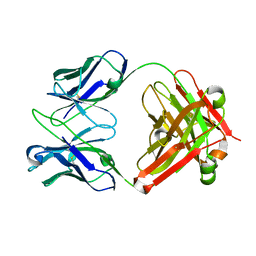 | | Crystal structure of Fab13B5 against HIV-1 capsid protein p24 | | Descriptor: | IMMUNOGLOBULIN HEAVY CHAIN, IMMUNOGLOBULIN LIGHT CHAIN | | Authors: | Monaco-Malbet, S, Berthet-Colominas, C, Novelli, A, Battai, N, Piga, N, Mallet, F, Cusack, S. | | Deposit date: | 2000-08-21 | | Release date: | 2000-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutual Conformational Adaptations in Antigen and Antibody Upon Complex Formation between an Fab and HIV-1 Capsid Protein P24
Structure, 8, 2000
|
|
