4U66
 
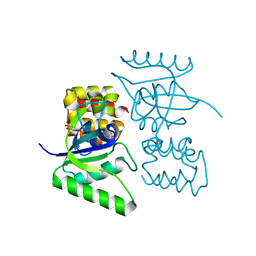 | |
4KNL
 
 | |
4U6I
 
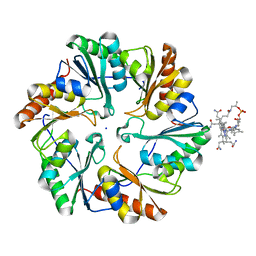 | | Crystal Structure of the EutL Microcompartment Shell Protein from Clostridium Perfringens Bound to Vitamin B12 | | Descriptor: | COBALAMIN, Ethanolamine utilization protein EutL, SODIUM ION | | Authors: | Thompson, M.C, Crowley, C.S, Kopstein, J.S, Yeates, T.O. | | Deposit date: | 2014-07-29 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a bacterial microcompartment shell protein bound to a cobalamin cofactor.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4U9W
 
 | | Crystal Structure of NatD bound to H4/H2A peptide and CoA | | Descriptor: | COENZYME A, GLYCEROL, Histone H4/H2A N-terminus, ... | | Authors: | Magin, R.S, Liszczak, G.P, Marmorstein, R. | | Deposit date: | 2014-08-06 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The Molecular Basis for Histone H4- and H2A-Specific Amino-Terminal Acetylation by NatD.
Structure, 23, 2015
|
|
4U6T
 
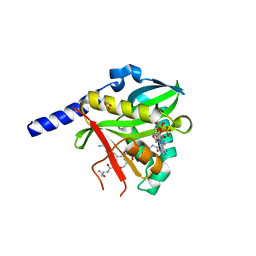
 | | Crystal structure of the Clostridium histolyticum colH collagenase polycystic kidney disease-like domain 2a at 1.76 Angstrom resolution | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ColH protein | | Authors: | Bauer, R, Janowska, K, Sakon, J, Matsushita, O, Latimer, E. | | Deposit date: | 2014-07-29 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structures of three polycystic kidney disease-like domains from Clostridium histolyticum collagenases ColG and ColH.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UAR
 
 | | Crystal structure of apo-CbbY from Rhodobacter sphaeroides | | Descriptor: | GLYCEROL, Protein CbbY | | Authors: | Bracher, A, Sharma, A, Starling-Windhof, A, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2014-08-11 | | Release date: | 2014-12-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Degradation of potent Rubisco inhibitor by selective sugar phosphatase.
Nat.Plants, 1, 2015
|
|
4UE7
 
 | | Thrombin in complex with 1-amidinopiperidine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2014-12-16 | | Release date: | 2015-08-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.129 Å) | | Cite: | Fragments Can Bind Either More Enthalpy or Entropy-Driven: Crystal Structures and Residual Hydration Pattern Suggest Why.
J.Med.Chem., 58, 2015
|
|
2RR4
 
 | | Complex structure of the zf-CW domain and the H3K4me3 peptide | | Descriptor: | Histone H3, ZINC ION, Zinc finger CW-type PWWP domain protein 1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-03-24 | | Release date: | 2010-09-15 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the zinc finger CW domain as a histone modification reader
Structure, 18, 2010
|
|
1S2Q
 
 | | Crystal structure of MAOB in complex with N-propargyl-1(R)-aminoindan (Rasagiline) | | Descriptor: | (1R)-N-(prop-2-en-1-yl)-2,3-dihydro-1H-inden-1-amine, Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Binda, C, Hubalek, F, Li, M, Herzig, Y, Sterling, J, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2004-01-09 | | Release date: | 2004-03-30 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structures of Monoamine Oxidase B in Complex with Four Inhibitors of the N-Propargylaminoindan Class.
J.Med.Chem., 47, 2004
|
|
4K3G
 
 | |
3MJ9
 
 | |
4U1V
 
 | | Crystal structure of the E. coli ribosome bound to linopristin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Noeske, J, Huang, J, Olivier, N.B, Giacobbe, R.A, Zambrowski, M, Cate, J.H.D. | | Deposit date: | 2014-07-16 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synergy of streptogramin antibiotics occurs independently of their effects on translation.
Antimicrob.Agents Chemother., 58, 2014
|
|
4U4Z
 
 | | Crystal structure of Phyllanthoside bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
3MIT
 
 | | Structure of Banana lectin-alpha-D-mannose complex | | Descriptor: | HEXANE-1,6-DIOL, Lectin, ZINC ION, ... | | Authors: | Sharma, A, Vijayan, M. | | Deposit date: | 2010-04-12 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Influence of glycosidic linkage on the nature of carbohydrate binding in beta-prism I fold lectins: an X-ray and molecular dynamics investigation on banana lectin-carbohydrate complexes
Glycobiology, 21, 2011
|
|
1J8F
 
 | |
4K6U
 
 | | Crystal structure of Ad37 fiber knob in complex with trivalent sialic acid inhibitor ME0386 | | Descriptor: | 3,3',3''-[nitrilotris(methanediyl-1H-1,2,3-triazole-4,1-diyl)]tripropan-1-ol, ACETATE ION, Fiber protein, ... | | Authors: | Stehle, T, Bauer, J. | | Deposit date: | 2013-04-16 | | Release date: | 2014-10-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Triazole linker-based trivalent sialic acid inhibitors of adenovirus type 37 infection of human corneal epithelial cells.
Org.Biomol.Chem., 13, 2015
|
|
4U9V
 
 | |
4UA3
 
 | |
1S5L
 
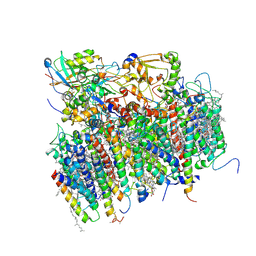 | | Architecture of the photosynthetic oxygen evolving center | | Descriptor: | 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, BETA-CAROTENE, BICARBONATE ION, ... | | Authors: | Ferreira, K.N, Iverson, T.M, Maghlaoui, K, Barber, J, Iwata, S. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-24 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Architecture of the Photosynthetic Oxygen-Evolving Center
Science, 303, 2004
|
|
4UEH
 
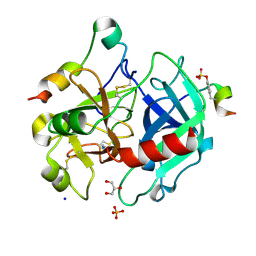 | | Thrombin in complex with benzamidine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BENZAMIDINE, GLYCEROL, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2014-12-17 | | Release date: | 2015-08-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Fragments Can Bind Either More Enthalpy or Entropy-Driven: Crystal Structures and Residual Hydration Pattern Suggest Why.
J.Med.Chem., 58, 2015
|
|
4KOM
 
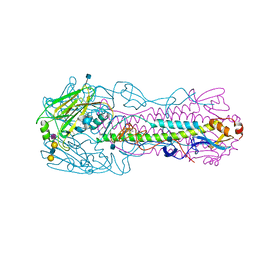 | | The structure of hemagglutinin from avian-origin H7N9 influenza virus in complex with avian receptor analog 3'SLNLN (NeuAcα2-3Galβ1-4GlcNAcβ1-3Galβ1-4Glc) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, Hemagglutinin HA2, ... | | Authors: | Shi, Y, Zhang, W, Wang, F, Qi, J, Song, H, Wu, Y, Gao, F, Zhang, Y, Fan, Z, Gong, W, Wang, D, Shu, Y, Wang, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-05-12 | | Release date: | 2013-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses.
Science, 342, 2013
|
|
2RFL
 
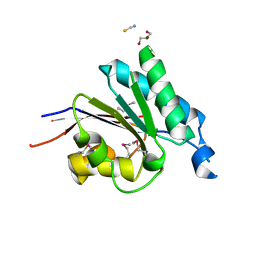 | | Crystal structure of the putative phosphohistidine phosphatase SixA from Agrobacterium tumefaciens | | Descriptor: | ACETIC ACID, GLYCEROL, Putative phosphohistidine phosphatase SixA, ... | | Authors: | Kim, Y, Binkowski, T, Xu, X, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-10-01 | | Release date: | 2007-10-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the Putative Phosphohistidine Phosphatase SixA from
Agrobacterium tumefaciens.
To be Published
|
|
4U3Q
 
 | |
4U4U
 
 | | Crystal structure of Lycorine bound to the yeast 80S ribosome | | Descriptor: | (1S,2S,12bS,12cS)-2,4,5,7,12b,12c-hexahydro-1H-[1,3]dioxolo[4,5-j]pyrrolo[3,2,1-de]phenanthridine-1,2-diol, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U3U
 
 | | Crystal structure of Cycloheximide bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-22 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
