1PJE
 
 | | Structure of the channel-forming trans-membrane domain of Virus protein "u"(Vpu) from HIV-1 | | Descriptor: | VPU protein | | Authors: | Park, S.H, Mrse, A.A, Nevzorov, A.A, Mesleh, M.F, Oblatt-Montal, M, Montal, M, Opella, S.J. | | Deposit date: | 2003-06-02 | | Release date: | 2003-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Three-dimensional structure of the channel-forming trans-membrane domain of Virus protein "u" (Vpu) from HIV-1
J.Mol.Biol., 333, 2003
|
|
2I3V
 
 | | Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor: Structure of G725C mutant | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Armstrong, N, Jasti, J, Beich-Frandsen, M, Gouaux, E. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Measurement of Conformational Changes accompanying Desensitization in an Ionotropic Glutamate Receptor.
Cell(Cambridge,Mass.), 127, 2006
|
|
3W5E
 
 | | Crystal structure of phosphodiesterase 4B in complex with compound 31e | | Descriptor: | CALCIUM ION, N-tert-butyl-2-{4-[(5,5-dioxido-2-phenyl-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-4-yl)amino]phenyl}acetamide, ZINC ION, ... | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2013-01-28 | | Release date: | 2013-05-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of the fused bicyclic 4-amino-2-phenylpyrimidine derivatives as novel and potent PDE4 inhibitors
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1F9H
 
 | | CRYSTAL STRUCTURE OF THE TERNARY COMPLEX OF E. COLI HPPK(R92A) WITH MGAMPCPP AND 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN AT 1.50 ANGSTROM RESOLUTION | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE, CHLORIDE ION, ... | | Authors: | Blaszczyk, J, Ji, X. | | Deposit date: | 2000-07-10 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Dynamic Roles of Arginine Residues 82 and 92 of Escherichia coli
6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase: Crystallographic
Studies
Biochemistry, 42, 2003
|
|
2E1Z
 
 | | Crystal structure of Salmonella typhimurium propionate kinase (TdcD) in complex with diadenosine tetraphosphate (Ap4A) obtained after co-crystallization with ATP | | Descriptor: | 1,2-ETHANEDIOL, BIS(ADENOSINE)-5'-TETRAPHOSPHATE, Propionate Kinase | | Authors: | Simanshu, D.K, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2006-11-04 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of Salmonella typhimurium propionate kinase and its complex with Ap4A: evidence for a novel Ap4A synthetic activity.
Proteins, 70, 2008
|
|
7M0Q
 
 | |
6JIC
 
 | | Identification and Characterization of a carboxypeptidase inhibitor from Lycium barbarum | | Descriptor: | WCI | | Authors: | Tan, W.L, Wong, K.H, Huang, J.Y, Tay, S.V, Wang, S.J. | | Deposit date: | 2019-02-20 | | Release date: | 2020-02-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification and characterization of a wolfberry carboxypeptidase inhibitor from Lycium barbarum.
Food Chem, 351, 2021
|
|
3D3P
 
 | |
2E20
 
 | | Crystal structure of Salmonella typhimurium propionate kinase (TdcD) in complex with diadenosine tetraphosphate (Ap4A) | | Descriptor: | 1,2-ETHANEDIOL, BIS(ADENOSINE)-5'-TETRAPHOSPHATE, Propionate Kinase | | Authors: | Simanshu, D.K, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2006-11-04 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Salmonella typhimurium propionate kinase and its complex with Ap4A: evidence for a novel Ap4A synthetic activity.
Proteins, 70, 2008
|
|
2E1Y
 
 | |
6MK5
 
 | |
6MZT
 
 | | Solution structure of alpha-KTx-6.21 (UroTx) from Urodacus yaschenkoi | | Descriptor: | Potassium channel toxin alpha-KTx 6.21 | | Authors: | Chin, Y.K.-Y, Luna-Ramirez, K, Anangi, R, King, G.F. | | Deposit date: | 2018-11-05 | | Release date: | 2020-03-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the potency and selectivity of Urotoxin, a potent Kv1 blocker from scorpion venom.
Biochem. Pharmacol., 174, 2020
|
|
8G5H
 
 | | Native GABA-A receptor from the mouse brain, ortho-alpha1-alpha3-beta2-gamma2 subtype, in complex with GABA, Zolpidem, and endogenous neurosteroid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2023-02-13 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
4MUW
 
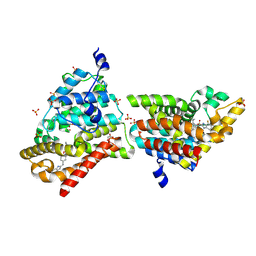 | | Crystal Structure of PDE10A with Novel Keto-Benzimidazole Inhibitor | | Descriptor: | 2-{4-[(6,7-difluoro-1H-benzimidazol-2-yl)amino]phenoxy}-N-methyl-3,4'-bipyridin-2'-amine, GLYCEROL, SULFATE ION, ... | | Authors: | Chmait, S, Jordan, S. | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Design, Optimization, and Biological Evaluation of Novel Keto-Benzimidazoles as Potent and Selective Inhibitors of Phosphodiesterase 10A (PDE10A).
J.Med.Chem., 56, 2013
|
|
1Q3E
 
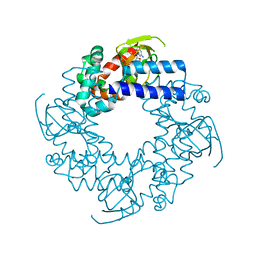 | | HCN2J 443-645 in the presence of cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Zagotta, W.N, Olivier, N.B, Black, K.D, Young, E.C, Olson, R, Gouaux, J.E. | | Deposit date: | 2003-07-29 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for modulation and agonist specificity of HCN pacemaker channels
Nature, 425, 2003
|
|
8G2U
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:control-apo-70S at 900ms | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8G34
 
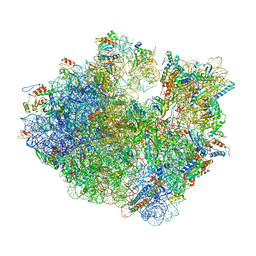 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:1st intermediate | | Descriptor: | 16S, 23S, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8G31
 
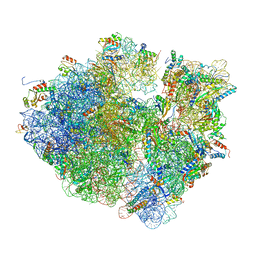 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:2nd Intermediate | | Descriptor: | 16S, 23S, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
6O3S
 
 | | NMR solution structure of Luffin P1 | | Descriptor: | Ribosome-inactivating protein luffin P1 | | Authors: | Rosengren, K.J, Payne, C. | | Deposit date: | 2019-02-27 | | Release date: | 2019-04-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | An Ancient Peptide Family Buried within Vicilin Precursors.
Acs Chem.Biol., 14, 2019
|
|
6O3Q
 
 | |
2JM4
 
 | |
6GNZ
 
 | |
4A3F
 
 | | RNA Polymerase II initial transcribing complex with a 6nt DNA-RNA hybrid and soaked with AMPCPP | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP*CP*TP*TP*DTP *TP*TP*CP*C BRU*GP*GP*TP*CP*AP*TP*T)-3', DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, ... | | Authors: | Cheung, A.C.M, Sainsbury, S, Cramer, P. | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of initial RNA polymerase II transcription.
EMBO J., 30, 2011
|
|
5WXN
 
 | | Structure of the LKB1 and 14-3-3 complex | | Descriptor: | 14-3-3 protein zeta/delta, Serine/threonine-protein kinase STK11 | | Authors: | Ding, S, Shi, Z.B. | | Deposit date: | 2017-01-08 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structure of the complex of phosphorylated liver kinase B1 and 14-3-3 zeta
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5WYO
 
 | | Solution structure of E.coli HdeA | | Descriptor: | Acid stress chaperone HdeA | | Authors: | Yang, C, Hu, Y, Jin, C. | | Deposit date: | 2017-01-14 | | Release date: | 2017-11-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Characterizations of the Interactions between Escherichia coli Periplasmic Chaperone HdeA and Its Native Substrates during Acid Stress
Biochemistry, 56, 2017
|
|
