2CG3
 
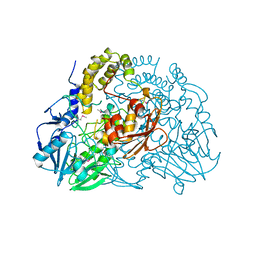 | | Crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa. | | Descriptor: | SDSA1, ZINC ION | | Authors: | Hagelueken, G, Adams, T.M, Wiehlmann, L, Widow, U, Kolmar, H, Tuemmler, B, Heinz, D.W, Schubert, W.-D. | | Deposit date: | 2006-02-27 | | Release date: | 2006-04-26 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of Sdsa1, an Alkylsulfatase from Pseudomonas Aeruginosa, Defines a Third Third Class of Sulfatases
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2B0J
 
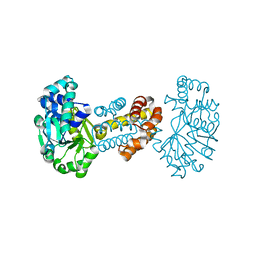 | | The crystal structure of the apoenzyme of the iron-sulfur-cluster-free hydrogenase (Hmd) | | Descriptor: | 5,10-methenyltetrahydromethanopterin hydrogenase | | Authors: | Pilak, O, Mamat, B, Vogt, S, Hagemeier, C.H, Thauer, R.K, Shima, S, Vonrhein, C, Warkentin, E, Ermler, U. | | Deposit date: | 2005-09-14 | | Release date: | 2006-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Crystal Structure of the Apoenzyme of the Iron-Sulphur Cluster-free Hydrogenase
J.Mol.Biol., 358, 2006
|
|
2ESY
 
 | | Structure and influence on stability and activity of the N-terminal propetide part of lung surfactant protein C | | Descriptor: | lung surfactant protein C | | Authors: | Li, J, Liepinsh, E, Almlen, A, Thyberg, J, Curstedt, T, Jornvall, H, Johansson, J. | | Deposit date: | 2005-10-27 | | Release date: | 2005-11-15 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure and influence on stability and activity of the N-terminal propeptide part of lung surfactant protein C
Febs J., 273, 2006
|
|
2FJB
 
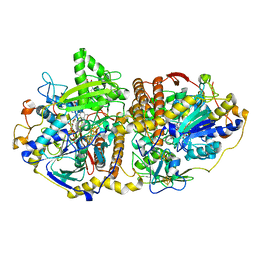 | | Adenosine-5'-phosphosulfate reductase im complex with products | | Descriptor: | (S)-10-((2S,3S,4R)-5-((S)-((S)-(((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHOXY)(HYDROXY)PHOSPHORYLOXY)(HYDROXY)PHOSPHORYLOXY)-2,3,4-TRIHYDROXYPENTYL)-7,8-DIMETHYL-2,4-DIOXO-2,3,4,4A-TETRAHYDROBENZO[G]PTERIDINE-5(10H)-SULFONIC ACID, ADENOSINE MONOPHOSPHATE, IRON/SULFUR CLUSTER, ... | | Authors: | Schiffer, A, Fritz, G, Kroneck, P.M, Ermler, U. | | Deposit date: | 2006-01-02 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reaction mechanism of the iron-sulfur flavoenzyme adenosine-5'-phosphosulfate reductase based on the structural characterization of different enzymatic states
Biochemistry, 45, 2006
|
|
3F47
 
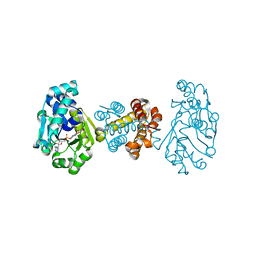 | | The Crystal Structure of [Fe]-Hydrogenase (Hmd) Holoenzyme from Methanocaldococcus jannaschii | | Descriptor: | 5'-O-[(S)-hydroxy{[2-hydroxy-3,5-dimethyl-6-(2-oxoethyl)pyridin-4-yl]oxy}phosphoryl]guanosine, 5,10-methenyltetrahydromethanopterin hydrogenase, CARBON MONOXIDE, ... | | Authors: | Hiromoto, T, Pilak, O, Warkentin, E, Thauer, R.K, Shima, S, Ermler, U. | | Deposit date: | 2008-10-31 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of C176A mutated [Fe]-hydrogenase suggests an acyl-iron ligation in the active site iron complex.
Febs Lett., 583, 2009
|
|
1JNR
 
 | | Structure of adenylylsulfate reductase from the hyperthermophilic Archaeoglobus fulgidus at 1.6 resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Fritz, G, Roth, A, Schiffer, A, Buechert, T, Bourenkov, G, Bartunik, H.D, Huber, H, Stetter, K.O, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2001-07-25 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of adenylylsulfate reductase from the hyperthermophilic Archaeoglobus fulgidus at 1.6-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JNZ
 
 | | Structure of adenylylsulfate reductase from the hyperthermophilic Archaeoglobus fulgidus at 1.6 resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, SULFITE ION, ... | | Authors: | Fritz, G, Roth, A, Schiffer, A, Buechert, T, Bourenkov, G, Bartunik, H.D, Huber, H, Stetter, K.O, Kroneck, P.M, Ermler, U. | | Deposit date: | 2001-07-26 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of adenylylsulfate reductase from the hyperthermophilic Archaeoglobus fulgidus at 1.6-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1K9B
 
 | | Crystal structure of the bifunctional soybean Bowman-Birk inhibitor at 0.28 nm resolution. Structural peculiarities in a folded protein conformation | | Descriptor: | BOWMAN-BIRK TYPE PROTEINASE INHIBITOR | | Authors: | Voss, R.H, Ermler, U, Essen, L.O, Wenzl, G, Kim, Y.M, Flecker, P. | | Deposit date: | 2001-10-29 | | Release date: | 2001-11-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the bifunctional soybean Bowman-Birk inhibitor at 0.28-nm resolution. Structural peculiarities in a folded protein conformation.
Eur.J.Biochem., 242, 1996
|
|
2JW6
 
 | | Solution structure of the DEAF1 MYND domain | | Descriptor: | Deformed epidermal autoregulatory factor 1 homolog, ZINC ION | | Authors: | Spadaccini, R, Perrin, H, Bottomley, M, Ansieu, S, Sattler, M. | | Deposit date: | 2007-10-08 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Retraction notice to "Structure and functional analysis of the MYND domain" [J. Mol. Biol. (2006) 358, 498-508].
J.Mol.Biol., 376, 2008
|
|
1SB3
 
 | | Structure of 4-hydroxybenzoyl-CoA reductase from Thauera aromatica | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-hydroxybenzoyl-CoA reductase alpha subunit, ... | | Authors: | Unciuleac, M, Warkentin, E, Page, C.C, Boll, M, Ermler, U. | | Deposit date: | 2004-02-10 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a Xanthine Oxidase-Related 4-Hydroxybenzoyl-CoA Reductase with an Additional [4Fe-4S] Cluster and an Inverted Electron Flow.
Structure, 12, 2004
|
|
2Y7S
 
 | |
2Y37
 
 | | The discovery of novel, potent and highly selective inhibitors of inducible nitric oxide synthase (iNOS) | | Descriptor: | 2-[(1R)-3-amino-1-phenyl-propoxy]-4-chloro-benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Cheshire, D.R, Andrews, G, Beaton, H.G, Birkinshaw, T.N, Boughton-Smith, N, Connolly, S, Cook, T.R, Cooper, A, Cooper, S.L, Cox, D, Dixon, J, Gensmantel, N, Hamley, P.J, Harrison, R, Hartopp, P, Kack, H, Luker, T, Mete, A, Millichip, I, Nicholls, D.J, Pimm, A.D, St-Gallay, S.A, Wallace, A.V. | | Deposit date: | 2010-12-19 | | Release date: | 2011-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Discovery of Novel, Potent and Highly Selective Inhibitors of Inducible Nitric Oxide Synthase (Inos).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2Y0F
 
 | | STRUCTURE OF GCPE (IspG) FROM THERMUS THERMOPHILUS HB27 | | Descriptor: | 4-HYDROXY-3-METHYLBUT-2-EN-1-YL DIPHOSPHATE SYNTHASE, IRON/SULFUR CLUSTER | | Authors: | Rekittke, I, Nonaka, T, Wiesner, J, Demmer, U, Warkentin, E, Jomaa, H, Ermler, U. | | Deposit date: | 2010-12-02 | | Release date: | 2011-01-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the E-1-Hydroxy-2-Methyl-But-2-Enyl-4-Diphosphate Synthase (Gcpe) from Thermus Thermophilus.
FEBS Lett., 585, 2011
|
|
5LAA
 
 | |
1SAU
 
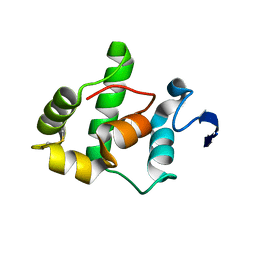 | | The Gamma subunit of the dissimilatory sulfite reductase (DsrC) from Archaeoglobus fulgidus at 1.1 A resolution | | Descriptor: | sulfite reductase, desulfoviridin-type subunit gamma | | Authors: | Mander, G.J, Weiss, M.S, Hedderich, R, Kahnt, J, Ermler, U, Warkentin, E. | | Deposit date: | 2004-02-09 | | Release date: | 2005-02-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Determination of a novel structure by a combination of long-wavelength sulfur phasing and radiation-damage-induced phasing.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1D6R
 
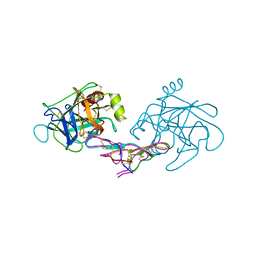 | | CRYSTAL STRUCTURE OF CANCER CHEMOPREVENTIVE BOWMAN-BIRK INHIBITOR IN TERNARY COMPLEX WITH BOVINE TRYPSIN AT 2.3 A RESOLUTION. STRUCTURAL BASIS OF JANUS-FACED SERINE PROTEASE INHIBITOR SPECIFICITY | | Descriptor: | BOWMAN-BIRK PROTEINASE INHIBITOR PRECURSOR, TRYPSINOGEN | | Authors: | Koepke, J, Ermler, U, Wenzl, G, Flecker, P. | | Deposit date: | 1999-10-15 | | Release date: | 2000-05-05 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of cancer chemopreventive Bowman-Birk inhibitor in ternary complex with bovine trypsin at 2.3 A resolution. Structural basis of Janus-faced serine protease inhibitor specificity.
J.Mol.Biol., 298, 2000
|
|
1D8B
 
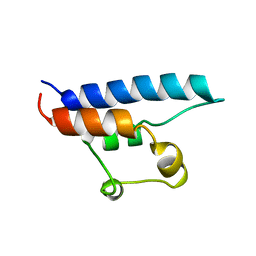 | | NMR STRUCTURE OF THE HRDC DOMAIN FROM SACCHAROMYCES CEREVISIAE RECQ HELICASE | | Descriptor: | SGS1 RECQ HELICASE | | Authors: | Liu, Z, Macias, M.J, Bottomley, M.J, Stier, G, Linge, J.P, Nilges, M, Bork, P, Sattler, M. | | Deposit date: | 1999-10-21 | | Release date: | 2000-01-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins.
Structure Fold.Des., 7, 1999
|
|
2ZNM
 
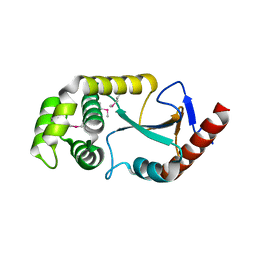 | | Oxidoreductase NmDsbA3 from Neisseria meningitidis | | Descriptor: | Thiol:disulfide interchange protein DsbA | | Authors: | Vivian, J.P, Scoullar, J, Robertson, A.L, Bottomley, S.P, Horne, J, Chin, Y, Velkov, T, Wielens, J, Thompson, P.E, Piek, S, Byres, E, Beddoe, T, Wilce, M.C.J, Kahler, C, Rossjohn, J, Scanlon, M.J. | | Deposit date: | 2008-04-30 | | Release date: | 2008-08-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Characterization of the Oxidoreductase NmDsbA3 from Neisseria meningitidis
J.Biol.Chem., 283, 2008
|
|
1U6K
 
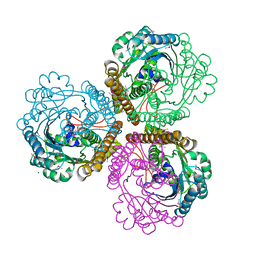 | | TLS refinement of the structure of Se-methionine labelled Coenzyme f420-dependent methylenetetrahydromethanopterin dehydrogenase (MTD) from Methanopyrus kandleri | | Descriptor: | F420-dependent methylenetetrahydromethanopterin dehydrogenase, MAGNESIUM ION | | Authors: | Warkentin, E, Hagemeier, C.H, Shima, S, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-07-30 | | Release date: | 2005-02-01 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structure of F420-dependent methylenetetrahydromethanopterin dehydrogenase: a crystallographic 'superstructure' of the selenomethionine-labelled protein crystal structure.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2YPV
 
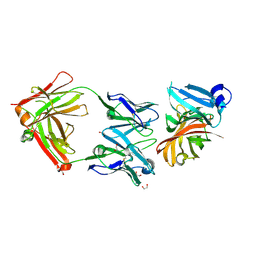 | | Crystal structure of the Meningococcal vaccine antigen factor H binding protein in complex with a bactericidal antibody | | Descriptor: | 1,2-ETHANEDIOL, FAB 12C1, LIPOPROTEIN | | Authors: | Malito, E, Veggi, D, Bottomley, M.J. | | Deposit date: | 2012-11-01 | | Release date: | 2013-02-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Defining a Protective Epitope on Factor H Binding Protein, a Key Meningococcal Virulence Factor and Vaccine Antigen.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1U6J
 
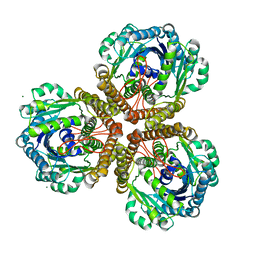 | | The Structure of native coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase at 2.4A resolution | | Descriptor: | F420-dependent methylenetetrahydromethanopterin dehydrogenase, MAGNESIUM ION | | Authors: | Warkentin, E, Hagemeier, C.H, Shima, S, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-07-30 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of F420-dependent methylenetetrahydromethanopterin dehydrogenase: a crystallographic 'superstructure' of the selenomethionine-labelled protein crystal structure.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1U6I
 
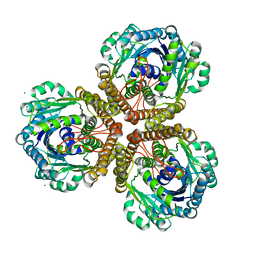 | | The Structure of native coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase at 2.2A resolution | | Descriptor: | F420-dependent methylenetetrahydromethanopterin dehydrogenase, MAGNESIUM ION | | Authors: | Warkentin, E, Hagemeier, C.H, Shima, S, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-07-30 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of F420-dependent methylenetetrahydromethanopterin dehydrogenase: a crystallographic 'superstructure' of the selenomethionine-labelled protein crystal structure.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1JAX
 
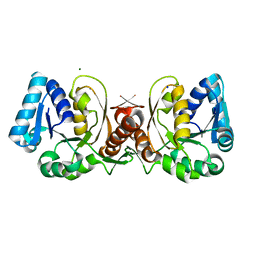 | | Structure of Coenzyme F420H2:NADP+ Oxidoreductase (FNO) | | Descriptor: | MAGNESIUM ION, SODIUM ION, conserved hypothetical protein | | Authors: | Warkentin, E, Mamat, B, Thauer, R, Ermler, U, Shima, S. | | Deposit date: | 2001-06-01 | | Release date: | 2001-12-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of F420H2:NADP+ oxidoreductase with and without its substrates bound.
EMBO J., 20, 2001
|
|
1K1G
 
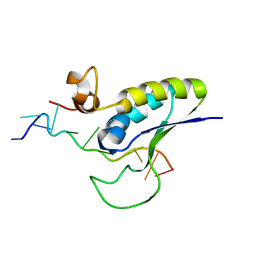 | | STRUCTURAL BASIS FOR RECOGNITION OF THE INTRON BRANCH SITE RNA BY SPLICING FACTOR 1 | | Descriptor: | 5'-R(*UP*AP*UP*AP*CP*UP*AP*AP*CP*AP*A)-3', SF1-Bo isoform | | Authors: | Liu, Z, Luyten, I, Bottomley, M.J, Messias, A.C, Houngninou-Molango, S, Sprangers, R, Zanier, K, Kramer, A, Sattler, M. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of the intron branch site RNA by splicing factor 1.
Science, 294, 2001
|
|
1JAY
 
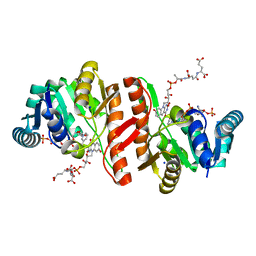 | | Structure of Coenzyme F420H2:NADP+ Oxidoreductase (FNO) with its substrates bound | | Descriptor: | COENZYME F420, Coenzyme F420H2:NADP+ Oxidoreductase (FNO), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Warkentin, E, Mamat, B, Thauer, R, Ermler, U, Shima, S. | | Deposit date: | 2001-06-01 | | Release date: | 2001-12-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of F420H2:NADP+ oxidoreductase with and without its substrates bound.
EMBO J., 20, 2001
|
|
