3QIZ
 
 | | Crystal Structure of BoNT/A LC complexed with Hydroxamate-based Inhibitor PT-2 | | Descriptor: | (2S,4R)-2-(2-{[3-(4-fluoro-3-methylphenyl)propyl](methyl)amino}ethyl)-4-(4-fluorophenyl)-N-hydroxy-4-methoxybutanamide, Botulinum neurotoxin type A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Thompson, A.A, Han, G.W, Stevens, R.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Three Novel Hydroxamate-Based Zinc Chelating Inhibitors of the Clostridium botulinum Serotype A Neurotoxin Light Chain Metalloprotease Reveals a Compact Binding Site Resulting from 60/70 Loop Flexibility.
Biochemistry, 50, 2011
|
|
3QJ0
 
 | | Crystal Structure of BoNT/A LC complexed with Hydroxamate-based Inhibitor PT-3 | | Descriptor: | (4R)-4-(4-chlorophenoxy)-1-[(4-chlorophenyl)sulfonyl]-N-hydroxy-L-prolinamide, 1,2-ETHANEDIOL, Botulinum neurotoxin type A, ... | | Authors: | Thompson, A.A, Han, G.W, Stevens, R.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural Characterization of Three Novel Hydroxamate-Based Zinc Chelating Inhibitors of the Clostridium botulinum Serotype A Neurotoxin Light Chain Metalloprotease Reveals a Compact Binding Site Resulting from 60/70 Loop Flexibility.
Biochemistry, 50, 2011
|
|
2FE8
 
 | | SARS coronavirus papain-like protease: structure of a viral deubiquitinating enzyme | | Descriptor: | BROMIDE ION, Replicase polyprotein 1ab, SULFATE ION, ... | | Authors: | Ratia, K, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2005-12-15 | | Release date: | 2006-03-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Severe acute respiratory syndrome coronavirus papain-like protease: Structure of a viral deubiquitinating enzyme
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1XYR
 
 | | Poliovirus 135S cell entry intermediate | | Descriptor: | Genome polyprotein, Coat protein VP1, Coat protein VP2, ... | | Authors: | Bubeck, D, Filman, D.J, Cheng, N, Steven, A.C, Hogle, J.M, Belnap, D.M. | | Deposit date: | 2004-11-10 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes.
J.Virol., 79, 2005
|
|
6HQ6
 
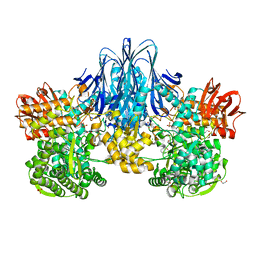 | | Bacterial beta-1,3-oligosaccharide phosphorylase from GH149 | | Descriptor: | 1,2-ETHANEDIOL, BICINE, Bacterial beta-1,3-oligosaccharide phosphorylase, ... | | Authors: | Kuhaudomlarp, S, Stevenson, C.E.M, Lawson, D.M, Field, R.A. | | Deposit date: | 2018-09-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of a GH149 beta-(1 → 3) glucan phosphorylase reveals a new surface oligosaccharide binding site and additional domains that are absent in the disaccharide-specific GH94 glucose-beta-(1 → 3)-glucose (laminaribiose) phosphorylase.
Proteins, 87, 2019
|
|
4QMD
 
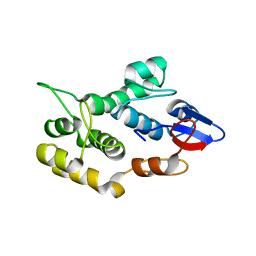 | | Crystal structure of human envoplakin plakin repeat domain | | Descriptor: | Envoplakin | | Authors: | Mohammed, F, Al-Jassar, C, White, S.A, Fogl, C, Jeeves, M, Knowles, T.J, Odinstova, E, Rodriguez-Zamora, P, Overduin, M, Chidgey, M. | | Deposit date: | 2014-06-16 | | Release date: | 2015-07-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Mechanism of intermediate filament recognition by plakin repeat domains revealed by envoplakin targeting of vimentin.
Nat Commun, 7, 2016
|
|
8WWR
 
 | |
4BD9
 
 | | Structure of the complex between SmCI and human carboxypeptidase A4 | | Descriptor: | CARBOXYPEPTIDASE A4, CARBOXYPEPTIDASE INHIBITOR SMCI, ZINC ION | | Authors: | Alonso-del-Ribero, M, Reytor, M.L, Trejo, S.A, Chavez, M.A, Aviles, F.X, Reverter, D. | | Deposit date: | 2012-10-05 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Noncanonical Mechanism of Carboxypeptidase Inhibition Revealed by the Crystal Structure of the Tri-Kunitz Smci in Complex with Human Cpa4.
Structure, 21, 2013
|
|
2ONX
 
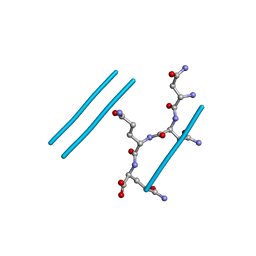 | | NNQQ peptide corresponding to residues 8-11 of yeast prion sup35 (alternate crystal form) | | Descriptor: | peptide corresponding to residues 8-11 of yeast prion sup35 | | Authors: | Sawaya, M.R, Sambashivan, S, Nelson, R, Ivanova, M, Sievers, S.A, Apostol, M.I, Thompson, M.J, Balbirnie, M, Wiltzius, J.J, McFarlane, H, Madsen, A.O, Riekel, C, Eisenberg, D. | | Deposit date: | 2007-01-24 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Atomic structures of amyloid cross-beta spines reveal varied steric zippers.
Nature, 447, 2007
|
|
4HON
 
 | |
4XKX
 
 | | Crystal structure of BACE1 in complex with 2-aminooxazoline 3-azaxanthene inhibitor 28 | | Descriptor: | (5S)-7-(2-fluoropyridin-3-yl)-3-(2-fluoropyridin-4-yl)spiro[chromeno[2,3-c]pyridine-5,4'-[1,3]oxazol]-2'-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-01-12 | | Release date: | 2015-02-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of 2-aminooxazoline 3-azaxanthenes as orally efficacious beta-secretase inhibitors for the potential treatment of Alzheimer's disease.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1KTK
 
 | | Complex of Streptococcal pyrogenic enterotoxin C (SpeC) with a human T cell receptor beta chain (Vbeta2.1) | | Descriptor: | Exotoxin type C, T-cell receptor beta chain | | Authors: | Sundberg, E.J, Li, H, Llera, A.S, McCormick, J.K, Tormo, J, Karjalainen, K, Schlievert, P.M, Mariuzza, R.A. | | Deposit date: | 2002-01-16 | | Release date: | 2002-06-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of two streptococcal superantigens bound to TCR beta chains reveal diversity in the architecture of T cell signaling complexes.
Structure, 10, 2002
|
|
5O2P
 
 | | p130Cas SH3 domain PTP-PEST peptide chimera | | Descriptor: | Breast cancer anti-estrogen resistance 1,Tyrosine-protein phosphatase non-receptor type 12 | | Authors: | Hexnerova, R, Veverka, V. | | Deposit date: | 2017-05-22 | | Release date: | 2017-09-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of CAS SH3 domain selectivity and regulation reveals new CAS interaction partners.
Sci Rep, 7, 2017
|
|
5O2Q
 
 | | p130Cas SH3 domain Vinculin peptide chimera | | Descriptor: | Breast cancer anti-estrogen resistance 1,Vinculin | | Authors: | Hexnerova, R, Veverka, V. | | Deposit date: | 2017-05-22 | | Release date: | 2017-09-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of CAS SH3 domain selectivity and regulation reveals new CAS interaction partners.
Sci Rep, 7, 2017
|
|
5O2M
 
 | | p130Cas SH3 domain | | Descriptor: | Breast cancer anti-estrogen resistance 1 | | Authors: | Hexnerova, R, Veverka, V. | | Deposit date: | 2017-05-22 | | Release date: | 2017-09-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of CAS SH3 domain selectivity and regulation reveals new CAS interaction partners.
Sci Rep, 7, 2017
|
|
5A1K
 
 | |
5A1M
 
 | | Crystal structure of calcium-bound human adseverin domain A3 | | Descriptor: | ADSEVERIN, CALCIUM ION | | Authors: | Chumnarnsilpa, S, Robinson, R.C, Grimes, J.M, Leyrat, C. | | Deposit date: | 2015-05-01 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Calcium-Controlled Conformational Choreography in the N-Terminal Half of Adseverin.
Nat.Commun., 6, 2015
|
|
7M1A
 
 | | SusE-like protein BT2857 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Galactose-binding like protein | | Authors: | Suits, M.D.L. | | Deposit date: | 2021-03-12 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Analysis of Two SusE-Like Enzymes From Bacteroides thetaiotaomicron Reveals a Potential Degradative Capacity for This Protein Family.
Front Microbiol, 12, 2021
|
|
7M1B
 
 | | SusE-like protein BT2857 | | Descriptor: | 1,2-ETHANEDIOL, Galactose-binding-like protein | | Authors: | Suits, M.D.L. | | Deposit date: | 2021-03-12 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Analysis of Two SusE-Like Enzymes From Bacteroides thetaiotaomicron Reveals a Potential Degradative Capacity for This Protein Family.
Front Microbiol, 12, 2021
|
|
1R44
 
 | | Crystal Structure of VanX | | Descriptor: | D-alanyl-D-alanine dipeptidase, ZINC ION | | Authors: | Pratt, S.D, Katz, L, Severin, J.M, Holzman, T, Park, C.H. | | Deposit date: | 2003-10-03 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure of VanX Reveals a Novel Amino-Dipeptidase Involved in Mediating Transposon-Based Vancomycin Resistance
Mol.Cell, 2, 1998
|
|
4DDX
 
 | |
4DDU
 
 | | Thermotoga maritima reverse gyrase, C2 FORM 1 | | Descriptor: | Reverse gyrase, ZINC ION | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2012-01-19 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Thermotoga maritima reverse gyrase: inferences for the mechanism of positive DNA supercoiling.
Nucleic Acids Res., 41, 2013
|
|
2FYG
 
 | | Crystal structure of NSP10 from Sars coronavirus | | Descriptor: | GLYCEROL, Replicase polyprotein 1ab, ZINC ION | | Authors: | Joseph, J.S, Saikatendu, K.S, Subramanian, V, Neuman, B.W, Brooun, A, Griffith, M, Moy, K, Yadav, M.K, Velasquez, J, Buchmeier, M.J, Stevens, R.C, Kuhn, P. | | Deposit date: | 2006-02-07 | | Release date: | 2006-08-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs.
J.Virol., 80, 2006
|
|
2CY6
 
 | | Crystal structure of ConM in complex with trehalose and maltose | | Descriptor: | CALCIUM ION, Lectin, MANGANESE (II) ION, ... | | Authors: | Cavada, B.S, Azevedo Jr, W.F, Delatorre, P, Rocha, B.A.M, Souza, E.P, Gadelha, C.A.A. | | Deposit date: | 2005-07-05 | | Release date: | 2006-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a lectin from Canavalia maritima (ConM) in complex with trehalose and maltose reveals relevant mutation in ConA-like lectins
J.Struct.Biol., 154, 2006
|
|
2CYF
 
 | | The Crystal Structure of Canavalia Maritima Lectin (ConM) in Complex with Trehalose and Maltose | | Descriptor: | CALCIUM ION, Lectin, MANGANESE (II) ION, ... | | Authors: | Delatorre, P, Rocha, B.A.M, Sousa, E.P, Gadelha, C.A.A, Azevedo Jr, W.F, Cavada, B.S. | | Deposit date: | 2005-07-06 | | Release date: | 2006-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a lectin from Canavalia maritima (ConM) in complex with trehalose and maltose reveals relevant mutation in ConA-like lectins
J.Struct.Biol., 154, 2006
|
|
