5Y6L
 
 | |
5UUO
 
 | | Crystal structure of SARO_2595 from Novosphingobium aromaticivorans | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase-like protein, ... | | Authors: | Bingman, C.A, Kontur, W.S, Olmsted, C.N, Fox, B.G, Donohue, T.J. | | Deposit date: | 2017-02-17 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Novosphingobium aromaticivoransuses a Nu-class glutathioneS-transferase as a glutathione lyase in breaking the beta-aryl ether bond of lignin.
J. Biol. Chem., 293, 2018
|
|
5UUN
 
 | | Crystal structure of SARO_2595 from Novosphingobium aromaticivorans | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione S-transferase-like protein | | Authors: | Bingman, C.A, Kontur, W.S, Olmsted, C.N, Fox, B.G, Donohue, T.J. | | Deposit date: | 2017-02-17 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novosphingobium aromaticivoransuses a Nu-class glutathioneS-transferase as a glutathione lyase in breaking the beta-aryl ether bond of lignin.
J. Biol. Chem., 293, 2018
|
|
5U56
 
 | | Structure of Francisella tularensis heterodimeric SspA (MglA-SspA) | | Descriptor: | GLYCEROL, Macrophage growth locus A, PENTAETHYLENE GLYCOL, ... | | Authors: | Cuthbert, B.J, Schumacher, M.A, Brennan, R.G. | | Deposit date: | 2016-12-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Dissection of the molecular circuitry controlling virulence in Francisella tularensis.
Genes Dev., 31, 2017
|
|
5U51
 
 | | Structure of Francisella tularensis heterodimeric SspA (MglA-SspA) in complex with ppGpp | | Descriptor: | GLYCEROL, GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Cuthbert, B.J, Schumacher, M.A, Brennan, R.G. | | Deposit date: | 2016-12-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dissection of the molecular circuitry controlling virulence in Francisella tularensis.
Genes Dev., 31, 2017
|
|
5O84
 
 | | Glutathione S-transferase Tau 23 (partially oxidized) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FORMIC ACID, ... | | Authors: | Young, D.R, Van Molle, I, Tossounian, M, Messens, J. | | Deposit date: | 2017-06-12 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Disulfide bond formation protects Arabidopsis thaliana glutathione transferase tau 23 from oxidative damage.
Biochim. Biophys. Acta, 1862, 2018
|
|
5O00
 
 | |
5NR1
 
 | | FzlA from C. crescentus | | Descriptor: | FtsZ-binding protein FzlA | | Authors: | Szwedziak, P, Lowe, J. | | Deposit date: | 2017-04-21 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | FzlA, an essential regulator of FtsZ filament curvature, controls constriction rate during Caulobacter division.
Mol. Microbiol., 107, 2018
|
|
5KEJ
 
 | | Crystallographic structure of the Tau class glutathione S-transferase MiGSTU in complex with S-hexyl-glutathione | | Descriptor: | DI(HYDROXYETHYL)ETHER, S-HEXYLGLUTATHIONE, Tau class glutathione S-transferase | | Authors: | Valenzuela-Chavira, I, Serrano-Posada, H, Lopez-Zavala, A, Hernandez-Paredes, J, Sotelo-Mundo, R. | | Deposit date: | 2016-06-09 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Insights into ligand binding to a glutathione S-transferase from mango: Structure, thermodynamics and kinetics.
Biochimie, 135, 2017
|
|
5JPO
 
 | |
5JCU
 
 | |
5HWL
 
 | | Human glutathione s-transferase Mu2 complexed with BDEA, monoclinic crystal form | | Descriptor: | GLUTATHIONE, Glutathione S-transferase Mu 2, N,N'-(butane-1,4-diyl)bis{2-[2,3-dichloro-4-(2-methylidenebutanoyl)phenoxy]acetamide} | | Authors: | Zhang, X, Wei, J, Wu, S, Zhang, H.P, Luo, M, Yang, X.L, Liao, F, Wang, D.Q. | | Deposit date: | 2016-01-29 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human glutathione s-transferase Mu2 complexed with BDEA, monoclinic crystal form
To Be Published
|
|
5HFK
 
 | | CRYSTAL STRUCTURE OF A GLUTATHIONE S-TRANSFERASE PROTEIN FROM ESCHERICHIA COLI OCh 157:H7 STR. SAKAI (ECs3186, TARGET EFI-507414) WITH BOUND GLUTATHIONE | | Descriptor: | Disulfide-bond oxidoreductase YfcG, GLUTATHIONE | | Authors: | Himmel, D.M, Toro, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-01-07 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | CRYSTAL STRUCTURE OF A GLUTATHIONE S-TRANSFERASE PROTEIN FROM ESCHERICHIA COLI OCh 157:H7 STR. SAKAI (ECs3186, TARGET EFI-507414) WITH BOUND GLUTATHIONE
TO BE PUBLISHED
|
|
5GST
 
 | | REACTION COORDINATE MOTION IN AN SNAR REACTION CATALYZED BY GLUTATHIONE TRANSFERASE | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE, SULFATE ION | | Authors: | Ji, X, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1993-07-20 | | Release date: | 1993-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Snapshots along the reaction coordinate of an SNAr reaction catalyzed by glutathione transferase.
Biochemistry, 32, 1993
|
|
5G5F
 
 | | Crystallographic structure of the Tau class glutathione S-transferase MiGSTU in complex with reduced glutathione. | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTATHIONE, TAU CLASS GLUTATHIONE S-TRANSFERASE | | Authors: | Valenzuela-Chavira, I, Serrano-Posada, H, Lopez-Zavala, A, Hernandez-Paredes, J, Sotelo-Mundo, R. | | Deposit date: | 2016-05-24 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights Into Ligand Binding to a Glutathione S-Transferase from Mango: Structure, Thermodynamics and Kinetics
Biochimie, 135, 2017
|
|
5G5E
 
 | | Crystallographic structure of the Tau class glutathione S-transferase MiGSTU from mango Mangifera indica L. | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, TAU CLASS GLUTATHIONE S-TRANSFERASE | | Authors: | Valenzuela-Chavira, I, Serrano-Posada, H, Lopez-Zavala, A, Hernandez-Paredes, J, Sotelo-Mundo, R. | | Deposit date: | 2016-05-24 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights Into Ligand Binding to a Glutathione S-Transferase from Mango: Structure, Thermodynamics and Kinetics
Biochimie, 135, 2017
|
|
5FWG
 
 | | TETRA-(5-FLUOROTRYPTOPHANYL)-GLUTATHIONE TRANSFERASE | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, TETRA-(5-FLUOROTRYPTOPHANYL)-GLUTATHIONE TRANSFERASE MU CLASS | | Authors: | Parsons, J.F, Xiao, G, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1997-11-08 | | Release date: | 1999-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymes harboring unnatural amino acids: mechanistic and structural analysis of the enhanced catalytic activity of a glutathione transferase containing 5-fluorotryptophan.
Biochemistry, 37, 1998
|
|
5FT3
 
 | | Aedes aegypti GSTe2 | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE EPSILON 2 | | Authors: | Yunta, C. | | Deposit date: | 2016-01-11 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.431 Å) | | Cite: | A Simple Test for the Determination of Ddt on Sprayed Surfaces
To be Published
|
|
5FHI
 
 | | Crystallographic structure of PsoE without Co | | Descriptor: | GLUTATHIONE, Glutathione S-transferase, putative | | Authors: | Hara, K, Hashimoto, H, Yamamoto, T, Tsunematsu, Y, Watanabe, K. | | Deposit date: | 2015-12-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Oxidative trans to cis Isomerization of Olefins in Polyketide Biosynthesis.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
5F8B
 
 | | Crystallographic Structure of PsoE with Co | | Descriptor: | (5~{S},8~{S},9~{R})-2-[(~{E})-hex-1-enyl]-8-methoxy-3-methyl-9-oxidanyl-8-(phenylcarbonyl)-1-oxa-7-azaspiro[4.4]non-2-ene-4,6-dione, COBALT (II) ION, GLUTATHIONE, ... | | Authors: | Hara, K, Hashimoto, H, Yamamoto, T, Tsunematsu, Y, Watanabe, K. | | Deposit date: | 2015-12-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Oxidative trans to cis Isomerization of Olefins in Polyketide Biosynthesis.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
5F0G
 
 | |
5F07
 
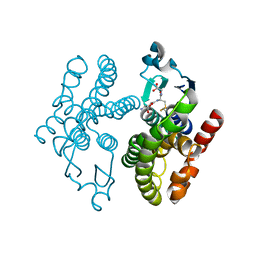 | | Crystal structure of glutathione transferase F8 from Populus trichocarpa | | Descriptor: | GLUTATHIONE, Putative glutathione S-transferase family protein | | Authors: | Didierjean, C, Rouhier, N, Pegeot, H, Gense, F. | | Deposit date: | 2015-11-27 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural plasticity among glutathione transferase Phi members: natural combination of catalytic residues confers dual biochemical activities.
FEBS J., 284, 2017
|
|
5F06
 
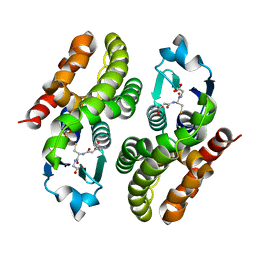 | | Crystal structure of glutathione transferase F7 from Populus trichocarpa | | Descriptor: | GLUTATHIONE, Glutathione S-transferase family protein, SULFATE ION | | Authors: | Didierjean, C, Rouhier, N, Pegeot, H, Gense, F. | | Deposit date: | 2015-11-27 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural plasticity among glutathione transferase Phi members: natural combination of catalytic residues confers dual biochemical activities.
FEBS J., 284, 2017
|
|
5F05
 
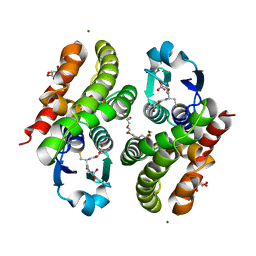 | | Crystal structure of glutathione transferase F5 from Populus trichocarpa | | Descriptor: | DODECAETHYLENE GLYCOL, GLUTATHIONE, GLYCEROL, ... | | Authors: | Didierjean, C, Rouhier, N, Pegeot, H, Gense, F. | | Deposit date: | 2015-11-27 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural plasticity among glutathione transferase Phi members: natural combination of catalytic residues confers dual biochemical activities.
FEBS J., 284, 2017
|
|
5EY6
 
 | |
