4MG7
 
 | | Crystal structure of hERa-LBD (Y537S) in complex with ferutinine | | Descriptor: | 1,2-ETHANEDIOL, Estrogen receptor, Nuclear receptor coactivator 1, ... | | Authors: | Delfosse, V, Grimaldi, M, Bourguet, W. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and functional profiling of environmental ligands for estrogen receptors.
Environ.Health Perspect., 122, 2014
|
|
4MGD
 
 | | Crystal structure of hERa-LBD (Y537S) in complex with HPTE | | Descriptor: | 1,2-ETHANEDIOL, 4,4'-(2,2,2-trichloroethane-1,1-diyl)diphenol, Estrogen receptor, ... | | Authors: | Delfosse, V, Grimaldi, M, Bourguet, W. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional profiling of environmental ligands for estrogen receptors.
Environ.Health Perspect., 122, 2014
|
|
4OY4
 
 | | calcium-free CaMPARI v0.2 | | Descriptor: | Chimera protein of Calmodulin, GPF-like protein EosFP, and Myosin light chain kinase, ... | | Authors: | Fosque, B.F, Schreiter, E.R. | | Deposit date: | 2014-02-10 | | Release date: | 2015-02-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Neural circuits. Labeling of active neural circuits in vivo with designed calcium integrators.
Science, 347, 2015
|
|
6BJV
 
 | | CIRV p19 protein in complex with siRNA | | Descriptor: | RNA (5'-R(P*CP*GP*UP*AP*CP*GP*CP*GP*GP*AP*AP*UP*AP*CP*UP*UP*CP*GP*AP*UP*U)-3'), RNA (5'-R(P*UP*CP*GP*AP*AP*GP*UP*AP*UP*UP*CP*CP*GP*CP*GP*UP*AP*CP*GP*UP*U)-3'), RNA silencing suppressor p19 | | Authors: | Foss, D.V, Schirle, N, Pezacki, J.P, Macrae, I.J. | | Deposit date: | 2017-11-07 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structural insights into interactions between viral suppressor of RNA silencing protein p19 mutants and small RNAs.
Febs Open Bio, 9, 2019
|
|
6BJG
 
 | | CIRV p19 mutant T111H in complex with siRNA | | Descriptor: | RNA (5'-R(P*CP*GP*UP*AP*CP*GP*CP*GP*GP*AP*AP*UP*AP*CP*UP*UP*CP*GP*AP*UP*U)-3'), RNA (5'-R(P*UP*CP*GP*AP*AP*GP*UP*AP*UP*UP*CP*CP*GP*CP*GP*UP*AP*CP*GP*UP*U)-3'), RNA silencing suppressor p19 | | Authors: | Foss, D.V, Schirle, N, Pezacki, J.P, Macrae, I.J. | | Deposit date: | 2017-11-06 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural insights into interactions between viral suppressor of RNA silencing protein p19 mutants and small RNAs.
Febs Open Bio, 9, 2019
|
|
6BJH
 
 | | CIRV p19 mutant T111S in complex with siRNA | | Descriptor: | RNA (5'-R(P*CP*GP*UP*AP*CP*GP*CP*GP*GP*AP*AP*UP*AP*CP*UP*UP*CP*GP*AP*UP*U)-3'), RNA (5'-R(P*UP*CP*GP*AP*AP*GP*UP*AP*UP*UP*CP*CP*GP*CP*GP*UP*AP*CP*GP*UP*U)-3'), RNA silencing suppressor p19 | | Authors: | Foss, D.V, Schirle, N.T, MacRae, I.J, Pezacki, J.P. | | Deposit date: | 2017-11-06 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural insights into interactions between viral suppressor of RNA silencing protein p19 mutants and small RNAs.
Febs Open Bio, 9, 2019
|
|
2C55
 
 | | Solution Structure of the Human Immunodeficiency Virus Type 1 p6 Protein | | Descriptor: | PROTEIN P6 | | Authors: | Fossen, T, Wray, V, Bruns, K, Rachmat, J, Henklein, P, Tessmer, U, Maczurek, A, Klinger, P, Schubert, U. | | Deposit date: | 2005-10-25 | | Release date: | 2005-11-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Human Immunodeficiency Virus Type 1 P6 Protein.
J.Biol.Chem., 280, 2005
|
|
5FMN
 
 | |
6P7E
 
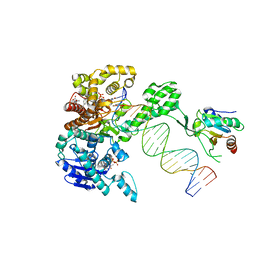 | | Structure of T7 DNA Polymerase Bound to a Primer/Template DNA and a Peptide that Mimics the C-terminal Tail of the Primase-Helicase | | Descriptor: | ASP-THR-ASP-PHE peptide, DNA (25-MER), DNA (5'-D(P*GP*GP*CP*AP*GP*GP*TP*GP*GP*TP*CP*TP*TP*GP*CP*CP*GP*GP*TP*GP*A)-3'), ... | | Authors: | Foster, B.M, Rosenberg, D, Salvo, H, Stephens, K.L, Bintz, B.J, Hammel, M, Ellenberger, T, Gainey, M.D, Wallen, J.R. | | Deposit date: | 2019-06-05 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Combined Solution and Crystal Methods Reveal the Electrostatic Tethers That Provide a Flexible Platform for Replication Activities in the Bacteriophage T7 Replisome.
Biochemistry, 58, 2019
|
|
5IQQ
 
 | |
7Q4K
 
 | | Erythromycin-stalled Escherichia coli 70S ribosome with streptococcal MsrDL nascent chain | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fostier, C.R, Ousalem, F, Soufari, H, Leroy, E.C, Ngo, S, Innis, A, Hashem, Y, Boel, G. | | Deposit date: | 2021-10-31 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Regulation of the macrolide resistance ABC-F translation factor MsrD.
Nat Commun, 14, 2023
|
|
1SQG
 
 | | The crystal structure of the E. coli Fmu apoenzyme at 1.65 A resolution | | Descriptor: | SUN protein | | Authors: | Foster, P.G, Nunes, C.R, Greene, P, Moustakas, D, Stroud, R.M. | | Deposit date: | 2004-03-18 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The First Structure of an RNA m5C Methyltransferase,
Fmu, Provides Insight into Catalytic Mechanism
and Specific Binding of RNA Substrate
Structure, 11, 2003
|
|
1SQF
 
 | | The crystal structure of E. coli Fmu binary complex with S-Adenosylmethionine at 2.1 A resolution | | Descriptor: | S-ADENOSYLMETHIONINE, SUN protein | | Authors: | Foster, P.G, Nunes, C.R, Greene, P, Moustakas, D, Stroud, R.M. | | Deposit date: | 2004-03-18 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The First Structure of an RNA m5C Methyltransferase,
Fmu, Provides Insight into Catalytic Mechanism
and Specific Binding of RNA Substrate
Structure, 11, 2003
|
|
1P6F
 
 | | Structure of the human natural cytotoxicity receptor NKp46 | | Descriptor: | natural cytotoxicity triggering receptor 1 | | Authors: | Foster, C.E, Colonna, M, Sun, P.D. | | Deposit date: | 2003-04-29 | | Release date: | 2003-12-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the human natural killer (NK) cell activating receptor NKp46 reveals structural relationship to other leukocyte receptor complex immunoreceptors.
J.Biol.Chem., 278, 2003
|
|
2HKL
 
 | | Crystal structure of Enterococcus faecium L,D-transpeptidase C442S mutant | | Descriptor: | L,D-TRANSPEPTIDASE, SULFATE ION | | Authors: | Delfosse, V, Hugonnet, J.-E, Magnet, S, Mainardi, J.-L, Arthur, M, Mayer, C. | | Deposit date: | 2006-07-05 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Enterococcus faecium L,D-transpeptidase C442S mutant
To be Published
|
|
6RC8
 
 | |
6RAO
 
 | |
9FQ4
 
 | |
9FOD
 
 | |
3UUD
 
 | | Crystal structure of hERa-LBD (Y537S) in complex with estradiol | | Descriptor: | 1,2-ETHANEDIOL, ESTRADIOL, Estrogen receptor, ... | | Authors: | Delfosse, V, Grimaldi, M, Bourguet, W. | | Deposit date: | 2011-11-28 | | Release date: | 2012-08-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and mechanistic insights into bisphenols action provide guidelines for risk assessment and discovery of bisphenol A substitutes.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6RGL
 
 | |
6RAP
 
 | |
6RBK
 
 | |
6RBN
 
 | |
2IST
 
 | |
