8GDI
 
 | | X-ray crystal structure of CYP124A1 from Mycobacterium Marinum in complex with 7-ketocholesterol | | Descriptor: | (3beta,8alpha,9beta)-3-hydroxycholest-5-en-7-one, Cytochrome P450 124A1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ghith, A, Bruning, J.B, Bell, S.G. | | Deposit date: | 2023-03-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The oxidation of cholesterol derivatives by the CYP124 and CYP142 enzymes from Mycobacterium marinum.
J.Steroid Biochem.Mol.Biol., 231, 2023
|
|
8GFC
 
 | |
8GFF
 
 | |
6WIZ
 
 | | Crystal structure of Fab 54-1G05 bound to H1 influenza hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 54-1G05 heavy chain, Fab 54-1G05 light chain, ... | | Authors: | Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-04-11 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Convergent Evolution in Breadth of Two VH6-1-Encoded Influenza Antibody Clonotypes from a Single Donor.
Cell Host Microbe, 28, 2020
|
|
8GFE
 
 | |
6WVV
 
 | | Plasmodium vivax M17 leucyl aminopeptidase | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, M17 leucyl aminopeptidase, ... | | Authors: | Malcolm, T.R, Drinkwater, N, McGowan, S. | | Deposit date: | 2020-05-07 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Active site metals mediate an oligomeric equilibrium in Plasmodium M17 aminopeptidases.
J.Biol.Chem., 296, 2020
|
|
8GFJ
 
 | |
8GFM
 
 | |
6WZC
 
 | |
8GFG
 
 | | Crystal structure of soluble lytic transglycosylase Cj0843 of Campylobacter jejuni in complex with Z7912 inhibitor | | Descriptor: | (3S,3aR,5S,6S,6aS)-2-oxohexahydro-2H-3,5-methanocyclopenta[b]furan-6-yl 2-acetamido-2-deoxy-beta-D-glucopyranoside, CITRIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2023-03-08 | | Release date: | 2023-05-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Exploring the inhibition of the soluble lytic transglycosylase Cj0843c of Campylobacter jejuni via targeting different sites with different scaffolds.
Protein Sci., 32, 2023
|
|
4ZX4
 
 | |
6WZZ
 
 | | GID4 in complex with VGLWKS peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, UNKNOWN ATOM OR ION, VGLWKS peptide | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-14 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of nonproline N-terminal residues by the Pro/N-degron pathway.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8GFH
 
 | |
8G1N
 
 | | Structure of Campylobacter concisus PglC I57M/Q175M Variant with modeled C-terminus | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, MAGNESIUM ION, N,N'-diacetylbacilliosaminyl-1-phosphate transferase, ... | | Authors: | Dodge, G.J, Ray, L.C, Das, D, Imperiali, B, Allen, K.N. | | Deposit date: | 2023-02-02 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Co-conserved sequence motifs are predictive of substrate specificity in a family of monotopic phosphoglycosyl transferases.
Protein Sci., 32, 2023
|
|
8IKD
 
 | |
4ZSJ
 
 | | MITOGEN ACTIVATED PROTEIN KINASE 7 IN COMPLEX WITH INHIBITOR | | Descriptor: | 3-amino-5-[(4-chloro-3-methylphenyl)amino]-N-(propan-2-yl)-1H-1,2,4-triazole-1-carboxamide, GLYCEROL, Mitogen-activated protein kinase 7 | | Authors: | Tucker, J, Ogg, D.J. | | Deposit date: | 2015-05-13 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5HRQ
 
 | | Insulin with proline analog HzP at position B28 in the R6 state | | Descriptor: | CHLORIDE ION, Insulin A-Chain, Insulin B-Chain, ... | | Authors: | Lieblich, S.A, Fang, K.Y, Cahn, J.K.B, Tirrell, D.A. | | Deposit date: | 2016-01-24 | | Release date: | 2017-01-25 | | Last modified: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | 4S-Hydroxylation of Insulin at ProB28 Accelerates Hexamer Dissociation and Delays Fibrillation.
J. Am. Chem. Soc., 139, 2017
|
|
5IUL
 
 | | Crystal structure of the DesK-DesR complex in the phosphotransfer state with high Mg2+ (150 mM) and BeF3 | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Trajtenberg, F, Imelio, J.A, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2016-03-18 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.153 Å) | | Cite: | Regulation of signaling directionality revealed by 3D snapshots of a kinase:regulator complex in action.
Elife, 5, 2016
|
|
6WPU
 
 | |
8IOF
 
 | |
5IVK
 
 | |
6WQ4
 
 | | Carbonic Anhydrase II Complexed with 2-((2-Cyanoethyl)(phenethyl)amino)-N-phenethyl-N-(4-sulfamoylphenethyl)acetamide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Andring, J.T, Combs, J.E, Lomelino, C, McKenna, R. | | Deposit date: | 2020-04-28 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Sulfonamide Inhibitors of Human Carbonic Anhydrases Designed through a Three-Tails Approach: Improving Ligand/Isoform Matching and Selectivity of Action.
J.Med.Chem., 63, 2020
|
|
9EY6
 
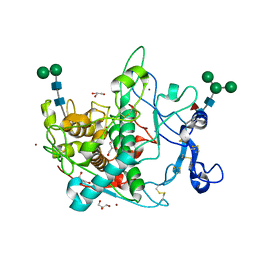 | | Crystal structure of human tyrosinase-related protein 1 (TYRP1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 5,6-dihydroxyindole-2-carboxylic acid oxidase, ... | | Authors: | Ng, Y.M, Soler-Lopez, M. | | Deposit date: | 2024-04-09 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.228 Å) | | Cite: | Interactions of Phenylalanine Derivatives with Human Tyrosinase: Lessons from Experimental and Theoretical tudies.
Chembiochem, 25, 2024
|
|
8ZJV
 
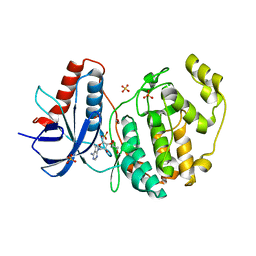 | | Crystal Structure of the ERK2 complexed with 5-Iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Yang, Y, Fu, L, Chen, R, Cheng, H, Sun, X. | | Deposit date: | 2024-05-15 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the ERK2 complexed with 5-Iodotubercidin
To Be Published
|
|
8IJG
 
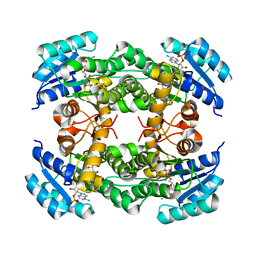 | | Crystal structure of alcohol dehydrogenase M5 from Burkholderia gladioli with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative short-chain dehydrogenases/reductase family protein | | Authors: | Han, X, Mei, Z.L, Liu, W.D, Sun, Z.T, Ma, J.A. | | Deposit date: | 2023-02-27 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of alcohol dehydrogenase from Burkholderia gladioli with NADP
To Be Published
|
|
