4HFB
 
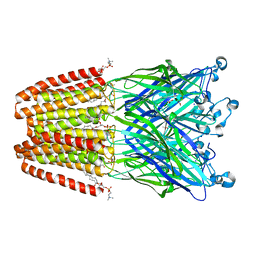 | | The GLIC pentameric Ligand-Gated Ion Channel F14'A ethanol-sensitive mutant (Apo) | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sauguet, L, Howard, R.J, Malherbe, L, Lee, U.S, Corringer, P.J, Harris, R.A, Delarue, M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for potentiation by alcohols and anaesthetics in a ligand-gated ion channel.
Nat Commun, 4, 2013
|
|
4GTM
 
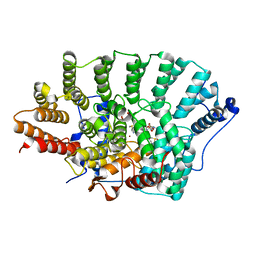 | | FTase in complex with BMS analogue 11 | | Descriptor: | 4-({(3R)-7-cyano-4-[(4-methoxyphenyl)sulfonyl]-1-[(1-methyl-1H-imidazol-5-yl)methyl]-2,3,4,5-tetrahydro-1H-1,4-benzodiazepin-3-yl}methyl)phenyl hexylcarbamate, DIMETHYL SULFOXIDE, FARNESYL DIPHOSPHATE, ... | | Authors: | Guo, Z, Stigter, E.A, Bon, R.S, Waldmann, H, Blankenfeldt, W, Goody, R.S. | | Deposit date: | 2012-08-28 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of Selective, Potent RabGGTase Inhibitors
J.Med.Chem., 55, 2012
|
|
6ZDV
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with Chromone 5d | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, ... | | Authors: | Verdon, G, Jespers, W, Azuaje, J, Majellaro, M, Keranen, H, Garcia-mera, X, Congreve, M, Deflorian, F, de Graaf, C, Zhukov, A, Dore, A, Mason, J, Aqvist, J, Cooke, R, Sotelo, E, Gutierrez-de-Teran, H. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | X-Ray Crystallography and Free Energy Calculations Reveal the Binding Mechanism of A 2A Adenosine Receptor Antagonists.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7LH7
 
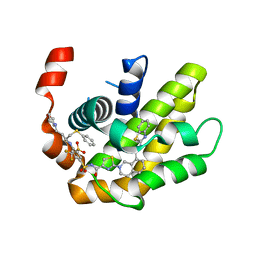 | | Crystal structure of BCL-XL in complex with a benzothiazole-based inhibitor | | Descriptor: | Bcl-2-like protein 1, N-(1,3-benzothiazol-2-yl)-2-(4-{[(4-{[(2R)-4-(morpholin-4-yl)-1-(phenylsulfanyl)butan-2-yl]amino}-3-[(trifluoromethyl)sulfonyl]phenyl)sulfonyl]carbamoyl}-1,3-thiazol-2-yl)-1,2,3,4-tetrahydroisoquinoline-8-carboxamide | | Authors: | Judge, R.A, Tao, Z. | | Deposit date: | 2021-01-21 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.409 Å) | | Cite: | Structure-Based Design of A-1293102, a Potent and Selective BCL-XL Inhibitor
ACS Medicinal Chemistry Letters, 12, 2021
|
|
3JBR
 
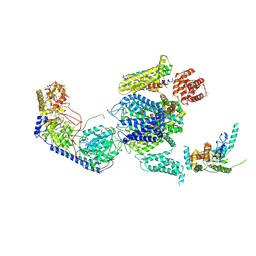 | | Cryo-EM structure of the rabbit voltage-gated calcium channel Cav1.1 complex at 4.2 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Voltage-dependent L-type calcium channel subunit alpha-1S, ... | | Authors: | Wu, J.P, Yan, Z, Yan, N. | | Deposit date: | 2015-09-29 | | Release date: | 2015-12-30 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the voltage-gated calcium channel Cav1.1 complex
Science, 350, 2015
|
|
4MQQ
 
 | | Mycobaterium tuberculosis transaminase BioA complexed with benzo[d]thiazole-2-carbohydrazide | | Descriptor: | (4-{[(E)-(1,3-benzothiazol-2-ylcarbonyl)diazenyl]methyl}-5-hydroxy-6-methylpyridin-3-yl)methyl dihydrogen phosphate, 1,2-ETHANEDIOL, Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, ... | | Authors: | Finzel, B.C, Dai, R. | | Deposit date: | 2013-09-16 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of Mycobacterium tuberculosis Transaminase BioA by Aryl Hydrazines and Hydrazides.
Chembiochem, 15, 2014
|
|
3JBY
 
 | | Cryo-electron microscopy structure of RAG Paired Complex (C2 symmetry) | | Descriptor: | '-D(P*GP*AP*TP*CP*TP*GP*GP*CP*CP*TP*GP*TP*CP*TP*TP*A)-3', 5'-D(P*CP*AP*CP*AP*GP*TP*GP*CP*TP*AP*CP*AP*GP*AP*C)-3', CALCIUM ION, ... | | Authors: | Ru, H, Chambers, M.G, Fu, T.-M, Tong, A.B, Liao, M, Wu, H. | | Deposit date: | 2015-10-22 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures.
Cell(Cambridge,Mass.), 163, 2015
|
|
4GWR
 
 | |
4GP8
 
 | | Structure of Recombinant Cytochrome ba3 Oxidase mutant Y133W+T231F from Thermus thermophilus | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Li, Y, Chen, Y, Stout, C.D. | | Deposit date: | 2012-08-20 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ligand Access to the Active Site in Thermus thermophilusba(3) and Bovine Heart aa(3) Cytochrome Oxidases.
Biochemistry, 52, 2013
|
|
4GYZ
 
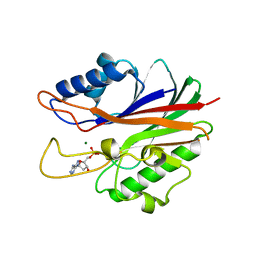 | | Mus Musculus Tdp2 Bound to dAMP and Mg2+ | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Schellenberg, M.J, Williams, R.S. | | Deposit date: | 2012-09-05 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.556 Å) | | Cite: | Mechanism of repair of 5'-topoisomerase II-DNA adducts by mammalian tyrosyl-DNA phosphodiesterase 2.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4KAX
 
 | | Crystal structure of the Grp1 PH domain in complex with Arf6-GTP | | Descriptor: | ADP-ribosylation factor 6, CITRIC ACID, Cytohesin-3, ... | | Authors: | Lambright, D.G, Malaby, A.W, van den Berg, B. | | Deposit date: | 2013-04-23 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for membrane recruitment and allosteric activation of cytohesin family Arf GTPase exchange factors.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3J7Z
 
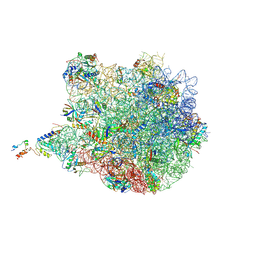 | | Structure of the E. coli 50S subunit with ErmCL nascent chain | | Descriptor: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Arenz, S, Meydan, S, Starosta, A.L, Berninghausen, O, Beckmann, R, Vazquez-Laslop, N, Wilson, D.N. | | Deposit date: | 2014-08-27 | | Release date: | 2014-10-22 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Drug Sensing by the Ribosome Induces Translational Arrest via Active Site Perturbation.
Mol.Cell, 56, 2014
|
|
4H0Y
 
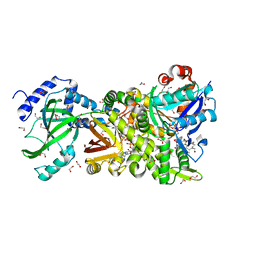 | | Crystal structure of NAD+-Ia(E380S)-actin complex | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Tsurumura, T, Oda, M, Nagahama, M, Tsuge, H. | | Deposit date: | 2012-09-10 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Arginine ADP-ribosylation mechanism based on structural snapshots of iota-toxin and actin complex
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3JT8
 
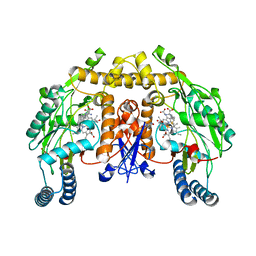 | | Structure of neuronal nitric oxide synthase heme domain complexed with N~5~-{3-[(1-methylethyl)sulfanyl]propanimidoyl}-L-ornithine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2009-09-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Heme-coordinating inhibitors of neuronal nitric oxide synthase. Iron-thioether coordination is stabilized by hydrophobic contacts without increased inhibitor potency.
J.Am.Chem.Soc., 132, 2010
|
|
4H3A
 
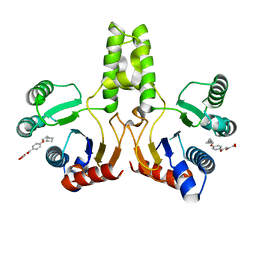 | |
3J8G
 
 | | Electron cryo-microscopy structure of EngA bound with the 50S ribosomal subunit | | Descriptor: | 23S rRNA, 50S ribosomal protein L1, 50S ribosomal protein L11, ... | | Authors: | Zhang, X, Yan, K, Zhang, Y, Li, N, Ma, C, Li, Z, Zhang, Y, Feng, B, Liu, J, Sun, Y, Xu, Y, Lei, J, Gao, N. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural insights into the function of a unique tandem GTPase EngA in bacterial ribosome assembly
Nucleic Acids Res., 2014
|
|
3JUY
 
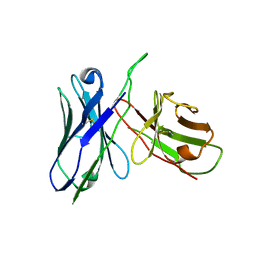 | |
3JWU
 
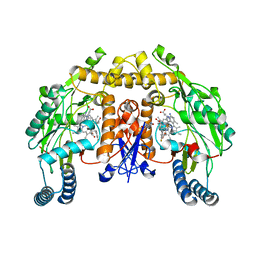 | | Structure of rat neuronal nitric oxide synthase R349A mutant heme domain in complex with N1-{(3'R,4'S)-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-fluorophenethyl)ethane-1,2-diamine tetrahydrochloride | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3R,4S)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}-N'-[2-(3-fluorophenyl)ethyl]ethane-1,2-diamine, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2009-09-18 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Unexpected binding modes of nitric oxide synthase inhibitors effective in the prevention of a cerebral palsy phenotype in an animal model.
J.Am.Chem.Soc., 132, 2010
|
|
7KSQ
 
 | | The Structure of the moss PSI-LHCI reveals the evolution of the LHCI antenna | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Riddle, R, Gorski, C, Toporik, H, Dobson, Z, Da, Z, Williams, D, Mazor, Y. | | Deposit date: | 2020-11-23 | | Release date: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The structure of the Physcomitrium patens photosystem I reveals a unique Lhca2 paralogue replacing Lhca4.
Nat.Plants, 8, 2022
|
|
4KGJ
 
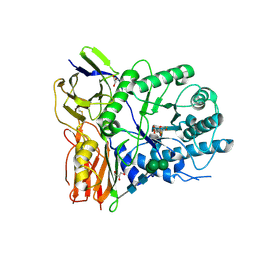 | | Crystal structure of human alpha-L-iduronidase complex with 5-fluoro-alpha-L-idopyranosyluronic acid fluoride | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-fluoro-alpha-L-idopyranosyluronic acid fluoride, ... | | Authors: | Bie, H, Yin, J, He, X, Kermode, A.R, Goddard-Borger, E.D, Withers, S.G, James, M.N.G. | | Deposit date: | 2013-04-29 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Insights into mucopolysaccharidosis I from the structure and action of alpha-L-iduronidase.
Nat.Chem.Biol., 9, 2013
|
|
3JX4
 
 | | Structure of rat neuronal nitric oxide synthase D597N/M336V mutant heme domain in complex with N1-{(3'R,4'S)-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-fluorophenethyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3R,4S)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}-N'-[2-(3-fluorophenyl)ethyl]ethane-1,2-diamine, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2009-09-18 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Unexpected binding modes of nitric oxide synthase inhibitors effective in the prevention of a cerebral palsy phenotype in an animal model.
J.Am.Chem.Soc., 132, 2010
|
|
4KH2
 
 | | Crystal structure of human alpha-L-iduronidase complex with 2-deoxy-2-fluoro-alpha-L-idopyranosyluronic acid fluoride | | Descriptor: | 2,6-anhydro-5-deoxy-5-fluoro-L-idonic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bie, H, Yin, J, He, X, Kermode, A.R, Goddard-Borger, E.D, Withers, S.G, James, M.N.G. | | Deposit date: | 2013-04-29 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Insights into mucopolysaccharidosis I from the structure and action of alpha-L-iduronidase.
Nat.Chem.Biol., 9, 2013
|
|
7KUX
 
 | | The Structure of the moss PSI-LHCI reveals the evolution of the LHCI antenna | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Riddle, R, Gorski, C, Toporik, H, Dobson, Z, Da, Z, Williams, D, Mazor, Y. | | Deposit date: | 2020-11-25 | | Release date: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The structure of the Physcomitrium patens photosystem I reveals a unique Lhca2 paralogue replacing Lhca4.
Nat.Plants, 8, 2022
|
|
3K09
 
 | | Crystal structure of the phosphorylation-site mutant S431D of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
4GXL
 
 | | The crystal structure of Galectin-8 C-CRD in complex with NDP52 | | Descriptor: | GLYCEROL, Galectin-8, Peptide from Calcium-binding and coiled-coil domain-containing protein 2 | | Authors: | Li, S, Wandel, M.P, Li, F, Liu, Z, He, C, Wu, J, Shi, Y, Randow, F. | | Deposit date: | 2012-09-04 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.023 Å) | | Cite: | Sterical hindrance promotes selectivity of the autophagy cargo receptor NDP52 for the danger receptor galectin-8 in antibacterial autophagy
Sci.Signal., 6, 2013
|
|
