5RAD
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD1B in complex with FM001568a | | Descriptor: | 2-[cyclohexyl(methylsulfonyl)amino]ethanamide, CHLORIDE ION, Lysine-specific demethylase 3B, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PanDDA analysis group deposition of Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
5RAY
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD1B in complex with FM001469a | | Descriptor: | 2,4-difluoro-6-[(3S)-pyrazolidin-3-yl]phenol, CHLORIDE ION, Lysine-specific demethylase 3B, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | PanDDA analysis group deposition of Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
5RAB
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD1B in complex with FM001726a | | Descriptor: | (2R,5S)-5-(4-chlorophenyl)oxolane-2-carbohydrazide, CHLORIDE ION, Lysine-specific demethylase 3B, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | PanDDA analysis group deposition of Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
5RAS
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD1B in complex with XS036302b | | Descriptor: | 2-(4-phenylpiperidin-1-yl)ethanoic acid, CHLORIDE ION, Lysine-specific demethylase 3B, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | PanDDA analysis group deposition of Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
5RAE
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD1B in complex with FM001558a | | Descriptor: | 3-[4-(4-hydroxyphenyl)phenyl]propanoic acid, CHLORIDE ION, Lysine-specific demethylase 3B, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | PanDDA analysis group deposition of Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
5RAV
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD1B in complex with FM001763a | | Descriptor: | CHLORIDE ION, Lysine-specific demethylase 3B, MANGANESE (II) ION, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | PanDDA analysis group deposition of Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
5R7X
 
 | | PanDDA analysis group deposition of ground-state model of Human JMJD1B | | Descriptor: | CHLORIDE ION, Lysine-specific demethylase 3B, MANGANESE (II) ION | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | PanDDA analysis group deposition of ground-state model for Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
5RAM
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD1B in complex with XS038544d | | Descriptor: | CHLORIDE ION, Lysine-specific demethylase 3B, MANGANESE (II) ION, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | PanDDA analysis group deposition of Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
5RB0
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD1B in complex with FM010020a | | Descriptor: | CHLORIDE ION, Lysine-specific demethylase 3B, MANGANESE (II) ION, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PanDDA analysis group deposition of Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
1AL9
 
 | |
5RAP
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD1B in complex with FM000707a | | Descriptor: | (3~{a}~{R},7~{a}~{R})-4-(4-methoxyphenyl)-2,3,3~{a},6,7,7~{a}-hexahydrothieno[3,2-c]pyridine, CHLORIDE ION, Lysine-specific demethylase 3B, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PanDDA analysis group deposition of Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
5RB6
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD1B in complex with FM001569a | | Descriptor: | 1-(4-chlorophenyl)-3-(2-methyl-1-oxidanyl-propan-2-yl)urea, CHLORIDE ION, Lysine-specific demethylase 3B, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | PanDDA analysis group deposition of Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
5RAA
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD1B in complex with FM009990a | | Descriptor: | CHLORIDE ION, Lysine-specific demethylase 3B, MANGANESE (II) ION, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | PanDDA analysis group deposition of Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
5RAQ
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD1B in complex with FM001577a | | Descriptor: | 2-[4-(2-methoxyphenyl)piperazin-1-yl]ethanenitrile, CHLORIDE ION, Lysine-specific demethylase 3B, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | PanDDA analysis group deposition of Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
5RB7
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD1B in complex with FM001648a | | Descriptor: | (1R,3S)-N-[(2H-1,3-benzodioxol-5-yl)methyl]-3-methylcyclopentan-1-amine, CHLORIDE ION, Lysine-specific demethylase 3B, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | PanDDA analysis group deposition of Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
5RAF
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD1B in complex with FM001559a | | Descriptor: | (5~{R})-3,4,4-trimethyl-5-(oxidanylamino)-1,3-thiazolidine-2-thione, CHLORIDE ION, Lysine-specific demethylase 3B, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | PanDDA analysis group deposition of Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
5RAU
 
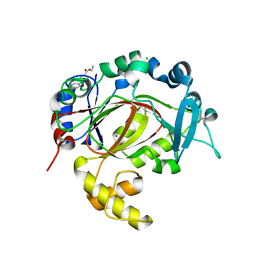 | | PanDDA analysis group deposition -- Crystal Structure of JMJD1B in complex with DA000165b | | Descriptor: | CHLORIDE ION, Lysine-specific demethylase 3B, MANGANESE (II) ION, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | PanDDA analysis group deposition of Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
1A40
 
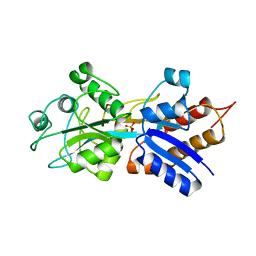 | | PHOSPHATE-BINDING PROTEIN WITH ALA 197 REPLACED WITH TRP | | Descriptor: | PHOSPHATE ION, PHOSPHATE-BINDING PERIPLASMIC PROTEIN PRECURSOR | | Authors: | Ledivina, P.S, Wang, Z, Tsai, A, Koehl, E, Quiocho, F.A. | | Deposit date: | 1998-02-10 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dominant role of local dipolar interactions in phosphate binding to a receptor cleft with an electronegative charge surface: equilibrium, kinetic, and crystallographic studies.
Protein Sci., 7, 1998
|
|
1IBA
 
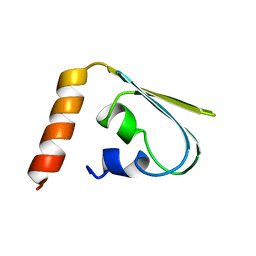 | | GLUCOSE PERMEASE (DOMAIN IIB), NMR, 11 STRUCTURES | | Descriptor: | GLUCOSE PERMEASE | | Authors: | Eberstadt, M, Grdadolnik, S.G, Gemmecker, G, Kessler, H, Buhr, A, Erni, B. | | Deposit date: | 1996-03-23 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the IIB domain of the glucose transporter of Escherichia coli.
Biochemistry, 35, 1996
|
|
1LFY
 
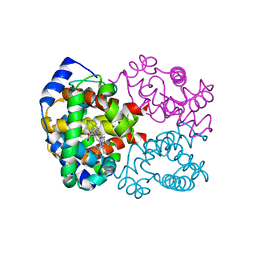 | | OXY HEMOGLOBIN (84% RELATIVE HUMIDITY) | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Biswal, B.K, Vijayan, M. | | Deposit date: | 2002-04-12 | | Release date: | 2002-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of human oxy- and deoxyhaemoglobin at different levels of humidity: variability in the T state.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1ZLP
 
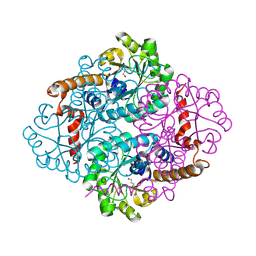 | | Petal death protein PSR132 with cysteine-linked glutaraldehyde forming a thiohemiacetal adduct | | Descriptor: | 5-HYDROXYPENTANAL, MAGNESIUM ION, petal death protein | | Authors: | Teplyakov, A, Liu, S, Lu, Z, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2005-05-08 | | Release date: | 2006-01-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Petal Death Protein from Carnation Flower.
Biochemistry, 44, 2005
|
|
1LFT
 
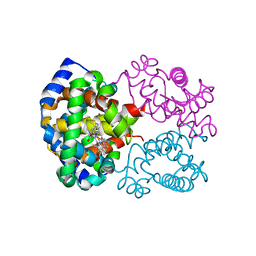 | | OXY HEMOGLOBIN (90% RELATIVE HUMIDITY) | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Biswal, B.K, Vijayan, M. | | Deposit date: | 2002-04-12 | | Release date: | 2002-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of human oxy- and deoxyhaemoglobin at different levels of humidity: variability in the T state.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1AMD
 
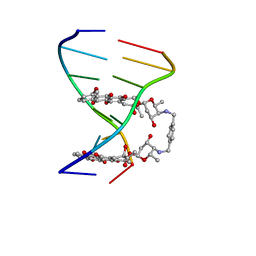 | |
2BOK
 
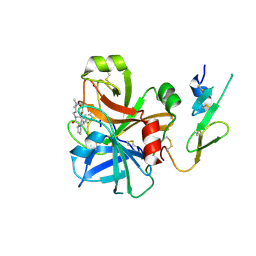 | | Factor Xa - cation | | Descriptor: | COAGULATION FACTOR X, SODIUM ION, [AMINO (4-{(3AS,4R,8AS,8BR)-1,3-DIOXO-2- [3-(TRIMETHYLAMMONIO) PROPYL]DECAHYDROPYRROLO[3,4-A] PYRROLIZIN-4-YL}PHENYL) METHYLENE]AMMONIUM | | Authors: | Morgenthaler, M, Schaerer, K, Paulini, R, Obst-Sander, U, Banner, D.W, Schlatter, D, Benz, J, Stihle, M, Diederich, F. | | Deposit date: | 2005-04-12 | | Release date: | 2005-06-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Quantification of Cation-Pi Interactions in Protein-Ligand Complexes: Crystal-Structure Analysis of Factor Xa Bound to a Quaternary Ammonium Ion Ligand
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1LFQ
 
 | | OXY HEMOGLOBIN (93% RELATIVE HUMIDITY) | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Biswal, B.K, Vijayan, M. | | Deposit date: | 2002-04-12 | | Release date: | 2002-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of human oxy- and deoxyhaemoglobin at different levels of humidity: variability in the T state.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
