2IO6
 
 | | Wee1 kinase complexed with inhibitor PD330961 | | Descriptor: | 9-HYDROXY-6-(3-HYDROXYPROPYL)-4-(2-METHOXYPHENYL)PYRROLO[3,4-C]CARBAZOLE-1,3(2H,6H)-DIONE, Wee1-like protein kinase | | Authors: | Squire, C.J, Dickson, J.M, Ivanovic, I, Baker, E.N. | | Deposit date: | 2006-10-10 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and structure-activity relationships of N-6 substituted analogues of 9-hydroxy-4-phenylpyrrolo[3,4-c]carbazole-1,3(2H,6H)-diones as inhibitors of Wee1 and Chk1 checkpoint kinases.
Eur.J.Med.Chem., 43, 2008
|
|
5ADI
 
 | | Structure of human nNOS R354A G357D mutant heme domain in complex with 7-(((5-((Methylamino)methyl)pyridin-3-yl)oxy)methyl)quinolin-2- amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[5-[(methylideneamino)methyl]pyridin-3-yl]oxymethyl]quinolin-2-amine, NITRIC OXIDE SYNTHASE, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-08-20 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phenyl Ether- and Aniline-Containing 2-Aminoquinolines as Potent and Selective Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
5ADG
 
 | | Structure of human nNOS R354A G357D mutant heme domain in complex with 7-((4-Chloro-3-((methylamino)methyl)phenoxy)methyl)quinolin-2- amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[4-chloranyl-3-(methylaminomethyl)phenoxy]methyl]quinolin-2-amine, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-08-20 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Phenyl Ether- and Aniline-Containing 2-Aminoquinolines as Potent and Selective Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
7L73
 
 | | Crystal structure of the first bromodomain (BD1) of human BRDT bound to ERK5-IN-1 | | Descriptor: | 11-cyclopentyl-2-({2-ethoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain testis-specific protein | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2020-12-25 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7LEJ
 
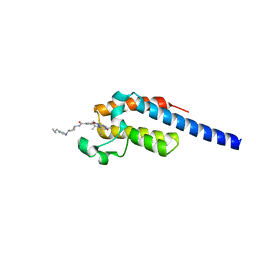 | | Crystal structure of the second bromodomain (BD2) of human BRDT bound to Volasertib | | Descriptor: | Bromodomain testis-specific protein, N-{trans-4-[4-(cyclopropylmethyl)piperazin-1-yl]cyclohexyl}-4-{[(7R)-7-ethyl-5-methyl-8-(1-methylethyl)-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxybenzamide | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7LEL
 
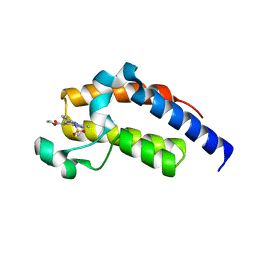 | |
7LEK
 
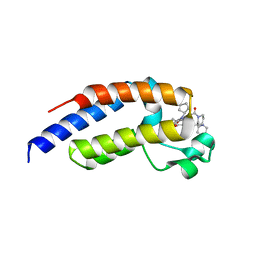 | | Crystal structure of the second bromodomain (BD2) of human BRDT bound to ERK5-IN-1 | | Descriptor: | 1,2-ETHANEDIOL, 11-cyclopentyl-2-({2-ethoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain testis-specific protein, ... | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
2K7J
 
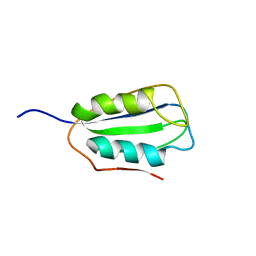 | | Human Acylphosphatase(AcPh) surface charge-optimized | | Descriptor: | Acylphosphatase-1 | | Authors: | Gribenko, A.V, Patel, M.M, Liu, J, McCallum, S.A, Wang, C, Makhatadze, G.I. | | Deposit date: | 2008-08-12 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Rational stabilization of enzymes by computational redesign of surface charge-charge interactions
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2JX3
 
 | |
2MB0
 
 | |
4MEO
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 2-methyl-quinoline ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, Vidler, L.R, Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Hoelder, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Novel Small-Molecule Inhibitors of BRD4 Using Structure-Based Virtual Screening.
J.Med.Chem., 56, 2013
|
|
4PCE
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with compound B13 | | Descriptor: | 1,2-ETHANEDIOL, 1-benzyl-2-ethyl-1,5,6,7-tetrahydro-4H-indol-4-one, Bromodomain-containing protein 4 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-04-15 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.293 Å) | | Cite: | Discovery of BRD4 bromodomain inhibitors by fragment-based high-throughput docking.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
6HK8
 
 | | Crystal structure of TEX12 delta-Ctip | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Testis-expressed protein 12 | | Authors: | Salmon, L.J, Davies, O.R. | | Deposit date: | 2018-09-06 | | Release date: | 2019-10-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.111 Å) | | Cite: | Synaptonemal complex central element extension through hierarchical assembly of SYCE2-TEX12 into alpha-fibres
To Be Published
|
|
4MEQ
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 5-methyl-triazolopyrimidine ligand | | Descriptor: | 1,2-ETHANEDIOL, 5-methyl-7-phenyl[1,2,4]triazolo[1,5-a]pyrimidin-2-amine, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, Vidler, L.R, Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Hoelder, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Discovery of Novel Small-Molecule Inhibitors of BRD4 Using Structure-Based Virtual Screening.
J.Med.Chem., 56, 2013
|
|
6F2U
 
 | | Potent and selective Aldo-Keto Reductase 1C3 (AKR1C3) inhibitors based on the benzoisoxazole moiety: application of a Bioisosteric Scaffold Hopping Approach to Flufenamic acid | | Descriptor: | 3-[(4-methoxyphenyl)methyl]-5-oxidanyl-~{N}-[3-(trifluoromethyl)phenyl]-1,2,3-triazole-4-carboxamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Goyal, P, Wahlgren, W.Y, Friemann, R. | | Deposit date: | 2017-11-27 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Potent and selective aldo-keto reductase 1C3 (AKR1C3) inhibitors based on the benzoisoxazole moiety: application of a bioisosteric scaffold hopping approach to flufenamic acid.
Eur J Med Chem, 150, 2018
|
|
2O36
 
 | |
8FL2
 
 | |
8FL4
 
 | |
8FL3
 
 | |
4O0F
 
 | | Crystal structure of the human L-asparaginase protein T219A mutant | | Descriptor: | GLYCINE, Isoaspartyl peptidase/L-asparaginase, SODIUM ION | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2013-12-13 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Elucidation of the Specific Function of the Conserved Threonine Triad Responsible for Human l-Asparaginase Autocleavage and Substrate Hydrolysis.
J.Mol.Biol., 426, 2014
|
|
4O0D
 
 | | Crystal structure of the human L-asparaginase protein T168S mutant | | Descriptor: | GLYCINE, Isoaspartyl peptidase/L-asparaginase, SODIUM ION | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2013-12-13 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Elucidation of the Specific Function of the Conserved Threonine Triad Responsible for Human l-Asparaginase Autocleavage and Substrate Hydrolysis.
J.Mol.Biol., 426, 2014
|
|
4O0H
 
 | |
6F78
 
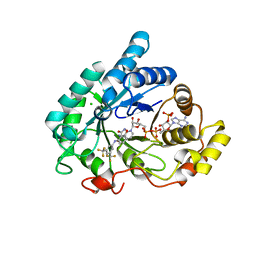 | | Potent and selective Aldo-Keto Reductase 1C3 (AKR1C3) inhibitors based on the benzoisoxazole moiety: Application of a Bioisosteric Scaffold Hopping Approach to Flufenamic acid | | Descriptor: | 4-[[3,5-bis(trifluoromethyl)phenyl]amino]-1,2-benzoxazol-3-one, Aldo-keto reductase family 1 member C3, CHLORIDE ION, ... | | Authors: | Goyal, P, Wahlgren, W.Y, Friemann, R. | | Deposit date: | 2017-12-07 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Potent and selective aldo-keto reductase 1C3 (AKR1C3) inhibitors based on the benzoisoxazole moiety: application of a bioisosteric scaffold hopping approach to flufenamic acid.
Eur J Med Chem, 150, 2018
|
|
4MEN
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 5-methyl-triazolopyrimidine ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N,5-dimethyl-N-(4-methylbenzyl)[1,2,4]triazolo[1,5-a]pyrimidin-7-amine | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, Vidler, L.R, Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Hoelder, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of Novel Small-Molecule Inhibitors of BRD4 Using Structure-Based Virtual Screening.
J.Med.Chem., 56, 2013
|
|
6H86
 
 | |
