5AKD
 
 | | MutS in complex with the N-terminal domain of MutL - crystal form 3 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (7.6 Å) | | Cite: | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
8F0V
 
 | | Lipocalin-like Milk protein-2 - E38A mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Milk protein, ZINC ION | | Authors: | Subramanian, R, KanagaVijayan, D. | | Deposit date: | 2022-11-04 | | Release date: | 2023-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Variability in phenylalanine side chain conformations facilitates broad substrate tolerance of fatty acid binding in cockroach milk proteins.
Plos One, 18, 2023
|
|
6WAO
 
 | |
8F0Y
 
 | | Lipocalin-like Milk protein-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Milk protein | | Authors: | Subramanian, R, KanagaVijayan, D, Shantakumar, R.P.S. | | Deposit date: | 2022-11-04 | | Release date: | 2023-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Variability in phenylalanine side chain conformations facilitates broad substrate tolerance of fatty acid binding in cockroach milk proteins.
Plos One, 18, 2023
|
|
5KV6
 
 | | Human cyclophilin A at 278K, Data set 8 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
6VR8
 
 | | Structure of a pseudomurein peptide ligase type E from Methanothermus fervidus | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Mur ligase middle domain protein, ... | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S, Subedi, B.P. | | Deposit date: | 2020-02-06 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterisation of methanogen pseudomurein cell wall peptide ligases homologous to bacterial MurE/F murein peptide ligases.
Microbiology (Reading, Engl.), 168, 2022
|
|
8INO
 
 | | Crystal structure of UGT74AN3 in complex UDP and PER | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[(3S,5S,8S,9S,10R,13R,14S,17R)-10,13-dimethyl-3,5,14-tris(oxidanyl)-2,3,4,6,7,8,9,11,12,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-17-yl]-2H-furan-5-one, Glycosyltransferase, ... | | Authors: | Long, F, Huang, W. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
5KWN
 
 | |
5AGU
 
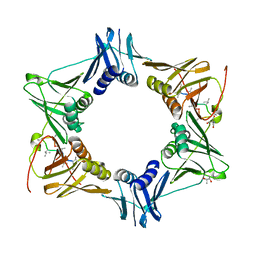 | | The sliding clamp of Mycobacterium tuberculosis in complex with a natural product. | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA POLYMERASE III SUBUNIT BETA, ... | | Authors: | Lukat, P, Kling, A, Heinz, D.W, Mueller, R. | | Deposit date: | 2015-02-03 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.173 Å) | | Cite: | Antibiotics. Targeting Dnan for Tuberculosis Therapy Using Novel Griselimycins.
Science, 348, 2015
|
|
4YRF
 
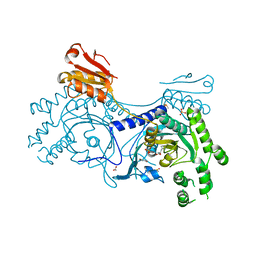 | | Crystal structure of T. cruzi Histidyl-tRNA synthetase in complex with 5-bromopyridin-2(1H)-one (Chem 148) | | Descriptor: | 1,2-ETHANEDIOL, 5-bromopyridin-2(1H)-one, DIMETHYL SULFOXIDE, ... | | Authors: | Koh, C.-Y, Hol, W.G.J. | | Deposit date: | 2015-03-15 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A binding hotspot in Trypanosoma cruzi histidyl-tRNA synthetase revealed by fragment-based crystallographic cocktail screens.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8FE7
 
 | |
8FE6
 
 | |
4YRN
 
 | |
5KZD
 
 | | N-acetylneuraminate lyase from methicillin-resistant Staphylococcus aureus with bound sialic acid alditol | | Descriptor: | (2~{S},4~{S},5~{R},6~{R},7~{S},8~{R})-5-acetamido-2,4,6,7,8,9-hexakis(oxidanyl)nonanoic acid, N-acetylneuraminate lyase | | Authors: | North, R.A, Watson, A.J.A, Pearce, F.G, Muscroft-Taylor, A.C, Friemann, R, Fairbanks, A.J, Dobson, R.C.J. | | Deposit date: | 2016-07-25 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.334 Å) | | Cite: | Structure and inhibition of N-acetylneuraminate lyase from methicillin-resistant Staphylococcus aureus.
FEBS Lett., 590, 2016
|
|
5L91
 
 | |
5LAN
 
 | |
8FUF
 
 | |
5L1R
 
 | | X-ray Structure of the Substrate-free Cytochrome P450 PntM | | Descriptor: | BICINE, PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
4YX1
 
 | | SpaO(SPOA2) | | Descriptor: | CALCIUM ION, Surface presentation of antigens protein SpaO | | Authors: | Notti, R.Q, Stebbins, C.E. | | Deposit date: | 2015-03-21 | | Release date: | 2015-06-03 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A common assembly module in injectisome and flagellar type III secretion sorting platforms.
Nat Commun, 6, 2015
|
|
5LBD
 
 | | Crystal structure of the N-domain of HMA6, a copper-transporting P-type ATPase | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, CHLORIDE ION, ... | | Authors: | Mayerhofer, H, Ravaud, S, Pebay-Peyroula, E. | | Deposit date: | 2016-06-15 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights into the Nucleotide-Binding Domains of the P1B-type ATPases HMA6 and HMA8 from Arabidopsis thaliana.
PLoS ONE, 11, 2016
|
|
5LBM
 
 | | The asymmetric tetrameric structure of the formaldehyde sensing transcriptional repressor FrmR from Escherichia coli | | Descriptor: | FORMYL GROUP, Transcriptional repressor FrmR | | Authors: | Bisson, C, Baker, P.J, Green, J, Chivers, P.T. | | Deposit date: | 2016-06-16 | | Release date: | 2016-12-21 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The mechanism of a formaldehyde-sensing transcriptional regulator.
Sci Rep, 6, 2016
|
|
5L21
 
 | |
4YX7
 
 | |
5L2J
 
 | | Crystal Structure of human CD1b in complex with C36-GMM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-[(2R,3R)-3-hydroxy-2-tetradecyldocosanoyl]-alpha-L-idopyranose, ACETATE ION, ... | | Authors: | Gras, S, Shahine, A, Le Nours, J, Rossjohn, J. | | Deposit date: | 2016-08-02 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | T cell receptor recognition of CD1b presenting a mycobacterial glycolipid.
Nat Commun, 7, 2016
|
|
4YNG
 
 | | Twinned pyruvate kinase from E. coli in the T-state | | Descriptor: | Pyruvate kinase I, SULFATE ION | | Authors: | Donovan, K.A, Dobson, R.C.J. | | Deposit date: | 2015-03-10 | | Release date: | 2015-03-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Grappling with anisotropic data, pseudo-merohedral twinning and pseudo-translational noncrystallographic symmetry: a case study involving pyruvate kinase.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
