4IWN
 
 | | Crystal structure of a putative methyltransferase CmoA in complex with a novel SAM derivative | | Descriptor: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, (4S)-2-METHYL-2,4-PENTANEDIOL, tRNA (cmo5U34)-methyltransferase | | Authors: | Aller, P, Lobley, C.M, Byrne, R.T, Antson, A.A, Waterman, D.G. | | Deposit date: | 2013-01-24 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | S-Adenosyl-S-carboxymethyl-L-homocysteine: a novel cofactor found in the putative tRNA-modifying enzyme CmoA.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2DNK
 
 | | Solution structure of RNA binding domain in Bruno-like 4 RNA binding protein | | Descriptor: | Bruno-like 4, RNA binding protein | | Authors: | Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-26 | | Release date: | 2006-10-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNA binding domain in Bruno-like 4 RNA binding protein
To be Published
|
|
4NPL
 
 | |
3ZHE
 
 | | Structure of the C. elegans SMG5-SMG7 complex | | Descriptor: | NONSENSE-MEDIATED MRNA DECAY PROTEIN, PROTEIN SMG-7 | | Authors: | Jonas, S, Weichenrieder, O, Izaurralde, E. | | Deposit date: | 2012-12-21 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | An Unusual Arrangement of Two 14-3-3-Like Domains in the Smg5-Smg7 Heterodimer is Required for Efficient Nonsense-Mediated Mrna Decay.
Genes Dev., 27, 2013
|
|
2OOE
 
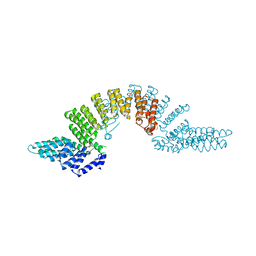 | | Crystal structure of HAT domain of murine CstF-77 | | Descriptor: | Cleavage stimulation factor 77 kDa subunit | | Authors: | Bai, Y, Auperin, T.C, Chou, C.-Y, Chang, G.-G, Manley, J.L, Tong, L. | | Deposit date: | 2007-01-25 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Murine CstF-77: Dimeric Association and Implications for Polyadenylation of mRNA Precursors.
Mol.Cell, 25, 2007
|
|
2OND
 
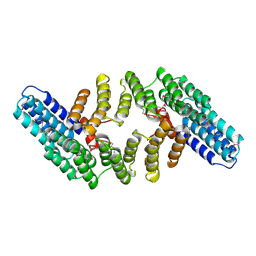 | | Crystal Structure of the HAT-C domain of murine CstF-77 | | Descriptor: | Cleavage stimulation factor 77 kDa subunit | | Authors: | Bai, Y, Auperin, T.C, Chou, C.-Y, Chang, G.-G, Manley, J.L, Tong, L. | | Deposit date: | 2007-01-23 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Murine CstF-77: Dimeric Association and Implications for Polyadenylation of mRNA Precursors.
Mol.Cell, 25, 2007
|
|
2YN2
 
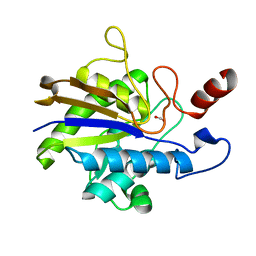 | | Huf protein - paralogue of the tau55 histidine phosphatase domain | | Descriptor: | FORMIC ACID, UNCHARACTERIZED PROTEIN YNL108C | | Authors: | Taylor, N.M.I, Glatt, S, Hennrich, M, von Scheven, G, Grotsch, H, Fernandez-Tornero, C, Rybin, V, Gavin, A.C, Kolb, P, Muller, C.W. | | Deposit date: | 2012-10-11 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Functional Characterization of a Phosphatase Domain within Yeast General Transcription Factor Tfiiic.
J.Biol.Chem., 288, 2013
|
|
4NPM
 
 | |
6BIC
 
 | | 2.25 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic inhibitor | | Descriptor: | (phenylmethyl) ~{N}-[(9~{S},12~{S},15~{S})-9-(hydroxymethyl)-12-(2-methylpropyl)-6,11,14-tris(oxidanylidene)-1,5,10,13,18,19-hexazabicyclo[15.2.1]icosa-17(20),18-dien-15-yl]carbamate, 3C-like protease | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Weerawarna, P.M, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Putative structural rearrangements associated with the interaction of macrocyclic inhibitors with norovirus 3CL protease.
Proteins, 87, 2019
|
|
6BIB
 
 | | 1.95 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic inhibitor | | Descriptor: | 3C-like protease, benzyl [(9S,12S,15S)-12-(cyclohexylmethyl)-9-(hydroxymethyl)-6,11,14-trioxo-1,5,10,13,18,19-hexaazabicyclo[15.2.1]icosa-17(20),18-dien-15-yl]carbamate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Weerawarna, P.M, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Putative structural rearrangements associated with the interaction of macrocyclic inhibitors with norovirus 3CL protease.
Proteins, 87, 2019
|
|
3KNP
 
 | |
4HEI
 
 | | 2A X-RAY STRUCTURE OF HPF from VIBRIO CHOLERAE | | Descriptor: | COBALT (III) ION, RIBOSOME HIBERNATION PROTEIN YHBH, THIOCYANATE ION, ... | | Authors: | De Bari, H, Berry, E.A. | | Deposit date: | 2012-10-03 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Vibrio cholerae ribosome hibernation promoting factor.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
6BID
 
 | | 1.15 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic inhibitor | | Descriptor: | 3C-like protease, benzyl [(8S,11S,14S)-11-(cyclohexylmethyl)-8-(hydroxymethyl)-5,10,13-trioxo-1,4,9,12,17,18-hexaazabicyclo[14.2.1]nonadeca-16(19),17-dien-14-yl]carbamate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Weerawarna, P.M, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Putative structural rearrangements associated with the interaction of macrocyclic inhibitors with norovirus 3CL protease.
Proteins, 87, 2019
|
|
2HA8
 
 | | Methyltransferase Domain of Human TAR (HIV-1) RNA binding protein 1 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, TAR (HIV-1) RNA loop binding protein | | Authors: | Min, J, Wu, H, Zeng, H, Loppnau, P, Battaile, K, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-12 | | Release date: | 2006-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of Methyltransferase Domain of Human TAR (HIV-1) RNA binding protein 1 in complex with S-adenosyl-L-homocystein.
To be Published
|
|
2EQS
 
 | | Solution structure of the S1 RNA binding domain of human ATP-dependent RNA helicase DHX8 | | Descriptor: | ATP-dependent RNA helicase DHX8 | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the S1 RNA binding domain of human ATP-dependent RNA helicase DHX8
To be Published
|
|
6GX9
 
 | | Crystal structure of the TNPO3 - CPSF6 RSLD complex | | Descriptor: | BENZAMIDINE, BICINE, Cleavage and polyadenylation specificity factor subunit 6, ... | | Authors: | Cherepanov, P, Cook, N. | | Deposit date: | 2018-06-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Differential role for phosphorylation in alternative polyadenylation function versus nuclear import of SR-like protein CPSF6.
Nucleic Acids Res., 47, 2019
|
|
2DK5
 
 | | Solution structure of Winged-Helix domain in RNA polymerase III 39KDa polypeptide | | Descriptor: | DNA-directed RNA polymerase III 39 kDa polypeptide | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-06 | | Release date: | 2006-10-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Winged-Helix domain in RNA polymerase III 39KDa polypeptide
To be Published
|
|
3EIP
 
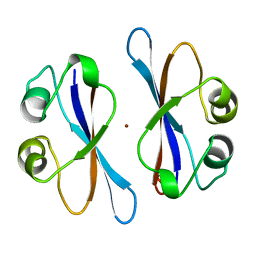 | | CRYSTAL STRUCTURE OF COLICIN E3 IMMUNITY PROTEIN: AN INHIBITOR TO A RIBOSOME-INACTIVATING RNASE | | Descriptor: | PROTEIN (COLICIN E3 IMMUNITY PROTEIN), ZINC ION | | Authors: | Li, C, Zhao, D, Djebli, A, Shoham, M. | | Deposit date: | 1999-03-29 | | Release date: | 1999-11-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of colicin E3 immunity protein: an inhibitor of a ribosome-inactivating RNase.
Structure Fold.Des., 7, 1999
|
|
1K6O
 
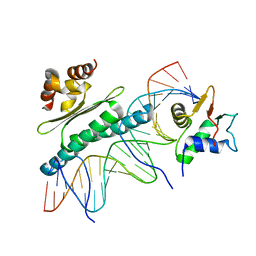 | | Crystal Structure of a Ternary SAP-1/SRF/c-fos SRE DNA Complex | | Descriptor: | 5'-D(*CP*AP*CP*AP*GP*GP*AP*TP*GP*TP*CP*CP*AP*TP*AP*TP*TP*AP*GP*GP*AP*CP*A)-3', 5'-D(*TP*GP*TP*CP*CP*TP*AP*AP*TP*AP*TP*GP*GP*AP*CP*AP*TP*CP*CP*TP*GP*TP*G)-3', ETS-domain protein ELK-4, ... | | Authors: | Mo, Y, Ho, W, Johnston, K, Marmorstein, R. | | Deposit date: | 2001-10-16 | | Release date: | 2002-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structure of a ternary SAP-1/SRF/c-fos SRE DNA complex.
J.Mol.Biol., 314, 2001
|
|
2KRC
 
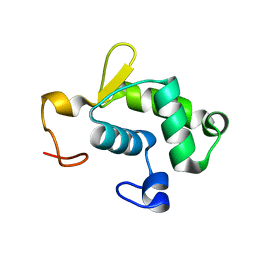 | | Solution structure of the N-terminal domain of Bacillus subtilis delta subunit of RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit delta | | Authors: | Motackova, V, Sanderova, H, Zidek, L, Novacek, J, Padrta, P, Svenkova, A, Jonak, J, Krasny, L, Sklenar, V. | | Deposit date: | 2009-12-16 | | Release date: | 2010-04-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of Bacillus subtilis delta subunit of RNA polymerase and its classification based on structural homologs
Proteins, 78, 2010
|
|
3BCA
 
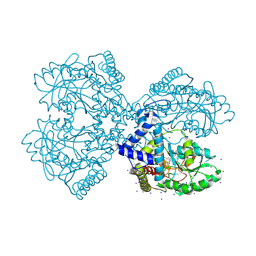 | |
1TBK
 
 | |
4K27
 
 | |
3BC8
 
 | | Crystal structure of mouse selenocysteine synthase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, O-phosphoseryl-tRNA(Sec) selenium transferase | | Authors: | Ganichkin, O.M, Wahl, M.C. | | Deposit date: | 2007-11-12 | | Release date: | 2007-12-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and catalytic mechanism of eukaryotic selenocysteine synthase.
J.Biol.Chem., 283, 2008
|
|
1VLR
 
 | |
