4R1S
 
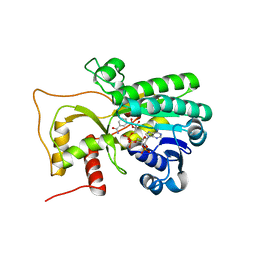 | | Crystal structure of Petunia hydrida cinnamoyl-CoA reductase | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, cinnamoyl CoA reductase | | Authors: | Noel, J.P, Louie, G.V, Bowman, M.E, Bomati, E.K. | | Deposit date: | 2014-08-07 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Studies of Cinnamoyl-CoA Reductase and Cinnamyl-Alcohol Dehydrogenase, Key Enzymes of Monolignol Biosynthesis.
Plant Cell, 26, 2014
|
|
5CTH
 
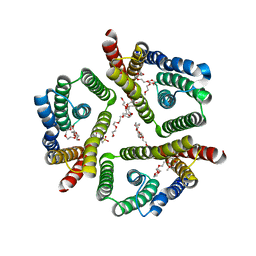 | | The 3.7 A resolution structure of a eukaryotic SWEET transporter | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Bidirectional sugar transporter SWEET2b, ... | | Authors: | Feng, L, Tao, Y, Perry, K. | | Deposit date: | 2015-07-24 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.69 Å) | | Cite: | Structure of a eukaryotic SWEET transporter in a homotrimeric complex.
Nature, 527, 2015
|
|
5CTG
 
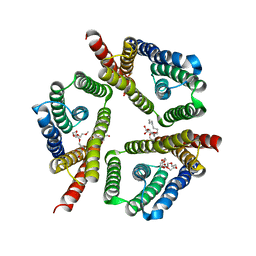 | | The 3.1 A resolution structure of a eukaryotic SWEET transporter | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Bidirectional sugar transporter SWEET2b, ... | | Authors: | Tao, Y, Perry, K, Feng, L. | | Deposit date: | 2015-07-24 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structure of a eukaryotic SWEET transporter in a homotrimeric complex.
Nature, 527, 2015
|
|
4QUK
 
 | |
2E0W
 
 | |
4R1U
 
 | | Crystal structure of Medicago truncatula cinnamoyl-CoA reductase | | Descriptor: | ACETATE ION, Cinnamoyl CoA reductase | | Authors: | Noel, J.P, Bomati, E.K, Louie, G.V, Bowman, M.E. | | Deposit date: | 2014-08-07 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural Studies of Cinnamoyl-CoA Reductase and Cinnamyl-Alcohol Dehydrogenase, Key Enzymes of Monolignol Biosynthesis.
Plant Cell, 26, 2014
|
|
1KY2
 
 | | GPPNHP-BOUND YPT7P AT 1.6 A RESOLUTION | | Descriptor: | GTP-BINDING PROTEIN YPT7P, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Constantinescu, A.-T, Rak, A, Scheidig, A.J. | | Deposit date: | 2002-02-02 | | Release date: | 2002-06-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rab-subfamily-specific regions of Ypt7p are structurally different from other RabGTPases.
Structure, 10, 2002
|
|
8GFT
 
 | | Hsp90 provides platform for CRaf dephosphorylation by PP5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, ... | | Authors: | Jaime-Garza, M, Nowotny, C.A, Coutandin, D, Wang, F, Tabios, M, Agard, D.A. | | Deposit date: | 2023-03-08 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Hsp90 provides a platform for kinase dephosphorylation by PP5.
Nat Commun, 14, 2023
|
|
2O3B
 
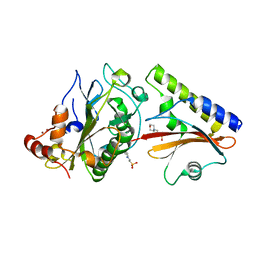 | | Crystal structure complex of Nuclease A (NucA) with intra-cellular inhibitor NuiA | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Ghosh, M, Meiss, G, Pingoud, A.M, London, R.E, Pedersen, L.C. | | Deposit date: | 2006-12-01 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The nuclease a-inhibitor complex is characterized by a novel metal ion bridge.
J.Biol.Chem., 282, 2007
|
|
2E0Y
 
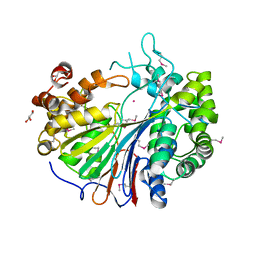 | |
4ON9
 
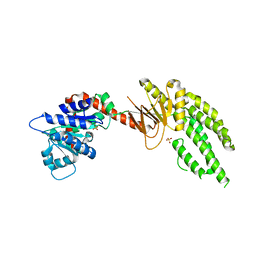 | | DECH box helicase domain | | Descriptor: | CHLORIDE ION, Probable ATP-dependent RNA helicase DDX58, SULFATE ION | | Authors: | Deimling, T, Witte, G, Hopfner, K.P. | | Deposit date: | 2014-01-28 | | Release date: | 2014-07-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal and solution structure of the human RIG-I SF2 domain
Acta Crystallogr.,Sect.F, 70, 2014
|
|
5D2J
 
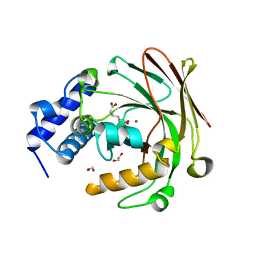 | |
5D2F
 
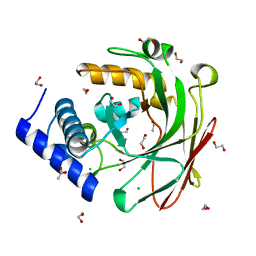 | | 4-oxalocrotonate decarboxylase from Pseudomonas putida G7 - apo form | | Descriptor: | 1,2-ETHANEDIOL, 4-oxalocrotonate decarboxylase NahK, ACETATE ION, ... | | Authors: | Guimaraes, S.L, Nagem, R.A.P. | | Deposit date: | 2015-08-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | Crystal Structures of Apo and Liganded 4-Oxalocrotonate Decarboxylase Uncover a Structural Basis for the Metal-Assisted Decarboxylation of a Vinylogous beta-Keto Acid.
Biochemistry, 55, 2016
|
|
5UFQ
 
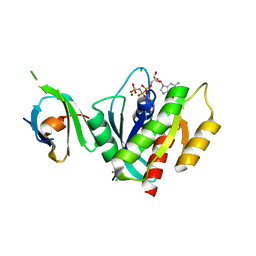 | | K-RasG12D(GNP)/R11.1.6 complex | | Descriptor: | CADMIUM ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Parker, J.A, Mattos, C. | | Deposit date: | 2017-01-05 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | An engineered protein antagonist of K-Ras/B-Raf interaction.
Sci Rep, 7, 2017
|
|
2J2C
 
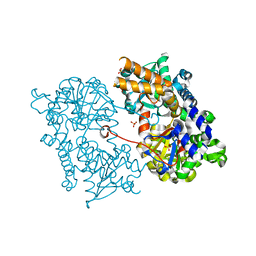 | | Crystal structure of Human Cytosolic 5'-Nucleotidase II (NT5C2, cN-II) | | Descriptor: | CYTOSOLIC PURINE 5'-NUCLEOTIDASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wallden, K, Stenmark, P, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Hogbom, M, Karlberg, T, Kotenyova, T, Loppnau, P, Magnusdottir, A, Nilsson-Ehle, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2006-08-16 | | Release date: | 2006-09-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Human Cytosolic 5'- Nucleotidase II: Insights Into Allosteric Regulation and Substrate Recognition
J.Biol.Chem., 282, 2007
|
|
5UFL
 
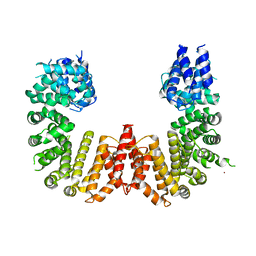 | | Crystal structure of a CIP2A core domain | | Descriptor: | Protein CIP2A, ZINC ION | | Authors: | Wang, Z, Wang, J, Rao, Z, Xu, W. | | Deposit date: | 2017-01-04 | | Release date: | 2017-02-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Oncoprotein CIP2A is stabilized via interaction with tumor suppressor PP2A/B56.
EMBO Rep., 18, 2017
|
|
3FTT
 
 | | Crystal Structure of the galactoside O-acetyltransferase from Staphylococcus aureus | | Descriptor: | Putative acetyltransferase SACOL2570 | | Authors: | Knapik, A.A, Shumilin, I.A, Cui, H, Xu, X, Chruszcz, M, Zimmerman, M.D, Cymborowski, M, Anderson, W.F, Savchenko, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-01-13 | | Release date: | 2009-03-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biophysical analysis of the putative acetyltransferase SACOL2570 from methicillin-resistant Staphylococcus aureus.
J.Struct.Funct.Genom., 14, 2013
|
|
3FSY
 
 | | Structure of tetrahydrodipicolinate N-succinyltransferase (Rv1201c;DapD) in complex with succinyl-CoA from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, MAGNESIUM ION, ... | | Authors: | Schuldt, L, Weyand, S, Kefala, G, Weiss, M.S. | | Deposit date: | 2009-01-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The three-dimensional Structure of a mycobacterial DapD provides insights into DapD diversity and reveals unexpected particulars about the enzymatic mechanism.
J.Mol.Biol., 389, 2009
|
|
3FRK
 
 | | X-ray structure of QdtB from T. thermosaccharolyticum in complex with a PLP:TDP-3-aminoquinovose aldimine | | Descriptor: | (2R,3R,4S,5S,6R)-3,5-dihydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, QdtB | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2009-01-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of QdtB, an aminotransferase required for the biosynthesis of dTDP-3-acetamido-3,6-dideoxy-alpha-D-glucose.
Biochemistry, 48, 2009
|
|
5D2I
 
 | |
4IPE
 
 | | Crystal structure of mitochondrial Hsp90 (TRAP1) with AMPPNP | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Partridge, J.R, Lavery, L.A, Agard, D.A. | | Deposit date: | 2013-01-09 | | Release date: | 2014-01-22 | | Last modified: | 2014-08-27 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Structural asymmetry in the closed state of mitochondrial Hsp90 (TRAP1) supports a two-step ATP hydrolysis mechanism.
Mol.Cell, 53, 2014
|
|
2E7Y
 
 | | High resolution structure of T. maritima tRNase Z | | Descriptor: | S-1,2-PROPANEDIOL, SULFATE ION, ZINC ION, ... | | Authors: | Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-15 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The structure of the flexible arm of Thermotoga maritima tRNase Z differs from those of homologous enzymes
Acta Crystallogr.,Sect.F, 63, 2007
|
|
1KSR
 
 | | THE REPEATING SEGMENTS OF THE F-ACTIN CROSS-LINKING GELATION FACTOR (ABP-120) HAVE AN IMMUNOGLOBULIN FOLD, NMR, 20 STRUCTURES | | Descriptor: | GELATION FACTOR | | Authors: | Fucini, P, Renner, C, Herberhold, C, Noegel, A.A, Holak, T.A. | | Deposit date: | 1997-02-07 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The repeating segments of the F-actin cross-linking gelation factor (ABP-120) have an immunoglobulin-like fold.
Nat.Struct.Biol., 4, 1997
|
|
4R1T
 
 | | Crystal structure of Petunia hydrida cinnamoyl-CoA reductase | | Descriptor: | cinnamoyl CoA reductase, molecular iodine | | Authors: | Noel, J.P, Louie, G.V, Bowman, M.E, Bomati, E.K. | | Deposit date: | 2014-08-07 | | Release date: | 2014-10-01 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Studies of Cinnamoyl-CoA Reductase and Cinnamyl-Alcohol Dehydrogenase, Key Enzymes of Monolignol Biosynthesis.
Plant Cell, 26, 2014
|
|
3AUU
 
 | | Crystal structure of Bacillus megaterium glucose dehydrogenase 4 in complex with D-glucose | | Descriptor: | Glucose 1-dehydrogenase 4, beta-D-glucopyranose | | Authors: | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, T. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided mutagenesis for the improvement of substrate specificity of Bacillus megaterium glucose 1-dehydrogenase IV
Febs J., 279, 2012
|
|
