4IR6
 
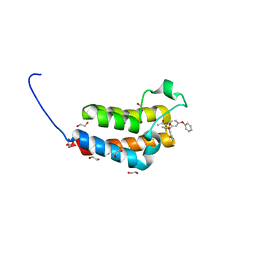 | | Crystal Structure of the bromodomain of human BAZ2B in complex with 1-{1-[2-(METHYLSULFONYL)PHENYL]-7-PHENOXYINDOLIZIN-3-YL}ETHANONE (GSK2838097A) | | Descriptor: | 1,2-ETHANEDIOL, 1-{1-[2-(methylsulfonyl)phenyl]-7-phenoxyindolizin-3-yl}ethanone, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, Chung, C.W, Drewry, D, Chen, P, Filippakopoulos, P, Fedorov, O, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and Characterization of GSK2801, a Selective Chemical Probe for the Bromodomains BAZ2A and BAZ2B.
J.Med.Chem., 59, 2016
|
|
2LZQ
 
 | |
2N68
 
 | |
2NTK
 
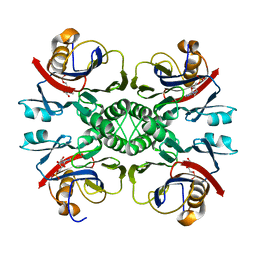 | | Crystal structure of PurO/IMP from Methanothermobacter thermoautotrophicus | | Descriptor: | IMP cyclohydrolase, INOSINIC ACID | | Authors: | Kang, Y.N, Tran, A, White, R.H, Ealick, S.E. | | Deposit date: | 2006-11-07 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A novel function for the N-terminal nucleophile hydrolase fold demonstrated by the structure of an archaeal inosine monophosphate cyclohydrolase.
Biochemistry, 46, 2007
|
|
7S7V
 
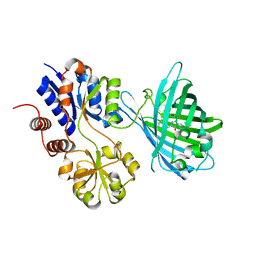 | | Crystal structure of iNicSnFR3a Fluorescent Nicotine Sensor | | Descriptor: | iNicSnFR 3.0 Fluorescent Nicotine Sensor | | Authors: | Fan, C, Shivange, A.V, Looger, L.L, Lester, H.A, Rees, D.C. | | Deposit date: | 2021-09-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Correction: Fluorescence activation mechanism and imaging of drug permeation with new sensors for smoking-cessation ligands.
Elife, 11, 2022
|
|
7S7Y
 
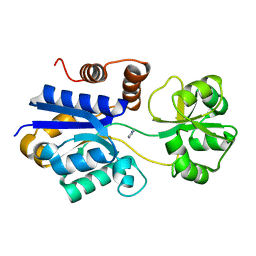 | | Crystal structure of iCytSnFR Cytisine Sensor precursor binding protein | | Descriptor: | IMIDAZOLE, iNicSnFR 4.0 Fluorescent Nicotine Sensor precursor binding protein | | Authors: | Fan, C, Nichols, N.L, Luebbert, L, Looger, L.L, Lester, H.A, Rees, D.C. | | Deposit date: | 2021-09-17 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure, Function, and Application of Bacterial ABC Transporters
Ph.D.Thesis,California Institute of Technology, 2020
|
|
7S7T
 
 | | iNicSnFR3a Nicotine Sensor comprising Periplasmic Binding sequence plus Fluorescent Sequence with varenicline bound | | Descriptor: | IODIDE ION, VARENICLINE, iNicSnFR 3.0 Fluorescent Nicotine Sensor | | Authors: | Fan, C, Shivange, A.V, Looger, L.L, Lester, H.A, Rees, D.C. | | Deposit date: | 2021-09-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Correction: Fluorescence activation mechanism and imaging of drug permeation with new sensors for smoking-cessation ligands.
Elife, 11, 2022
|
|
7S7U
 
 | | Crystal structure of iNicSnFR3a Fluorescent Nicotine Sensor with nicotine bound | | Descriptor: | iNicSnFR 3.0 Fluorescent Nicotine Sensor | | Authors: | Fan, C, Shivange, A.V, Looger, L.L, Lester, H.A, Rees, D.C. | | Deposit date: | 2021-09-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Correction: Fluorescence activation mechanism and imaging of drug permeation with new sensors for smoking-cessation ligands.
Elife, 11, 2022
|
|
7S7X
 
 | | Crystal structure of iCytSnFR Cytisine Sensor precursor binding protein with varenicline bound | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, DI(HYDROXYETHYL)ETHER, VARENICLINE, ... | | Authors: | Fan, C, Nichols, N.L, Luebbert, L, Looger, L.L, Lester, H.A, Rees, D.C. | | Deposit date: | 2021-09-17 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure, Function, and Application of Bacterial ABC Transporters
Ph.D.Thesis,California Institute of Technology, 2020
|
|
6L2C
 
 | | Crystal structure of Aspergillus fumigatus mitochondrial acetyl-CoA acetyltransferase in complex with CoA | | Descriptor: | Acetyl-CoA-acetyltransferase, putative, COENZYME A | | Authors: | Zhang, Y, Wei, W, Raimi, O.G, Ferenbach, A.T, Fang, W. | | Deposit date: | 2019-10-03 | | Release date: | 2020-02-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Aspergillus fumigatus Mitochondrial Acetyl Coenzyme A Acetyltransferase as an Antifungal Target.
Appl.Environ.Microbiol., 86, 2020
|
|
6LEM
 
 | | Structure of E. coli beta-glucuronidase complex with C6-nonyl uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-2-nonyl-4,5-bis(oxidanyl)piperidine-3-carboxylic acid, Beta-D-glucuronidase | | Authors: | Lin, H.-Y, Kuo, Y.-H, Lin, C.-H. | | Deposit date: | 2019-11-25 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.188 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
6LEG
 
 | | Structure of E. coli beta-glucuronidase complex with uronic isofagomine | | Descriptor: | (3S,4R,5R)-4,5-dihydroxypiperidine-3-carboxylic acid, Beta-D-glucuronidase | | Authors: | Lin, H.-Y, Kuo, Y.-H, Lin, C.-H. | | Deposit date: | 2019-11-25 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
6LEJ
 
 | | Structure of E. coli beta-glucuronidase complex with C6-propyl uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-4,5-bis(oxidanyl)-2-propyl-piperidine-3-carboxylic acid, Beta-D-glucuronidase | | Authors: | Lin, H.-Y, Kuo, Y.-H, Lin, C.-H. | | Deposit date: | 2019-11-25 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.617 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
6LEL
 
 | | Structure of E. coli beta-glucuronidase complex with C6-hexyl uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-2-hexyl-4,5-bis(oxidanyl)piperidine-3-carboxylic acid, Beta-D-glucuronidase | | Authors: | Lin, H.-Y, Kuo, Y.-H, Lin, C.-H. | | Deposit date: | 2019-11-25 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
4WFD
 
 | | Structure of the Rrp6-Rrp47-Mtr4 interaction | | Descriptor: | ATP-dependent RNA helicase DOB1, Exosome complex exonuclease RRP6, Exosome complex protein LRP1, ... | | Authors: | Schuch, B, Conti, E. | | Deposit date: | 2014-09-14 | | Release date: | 2014-10-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase.
Embo J., 33, 2014
|
|
4WFC
 
 | | Structure of the Rrp6-Rrp47 interaction | | Descriptor: | Exosome complex exonuclease RRP6, Exosome complex protein LRP1, SULFATE ION | | Authors: | Schuch, B, Conti, E. | | Deposit date: | 2014-09-14 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase.
Embo J., 33, 2014
|
|
6D39
 
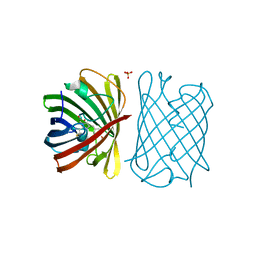 | |
6D38
 
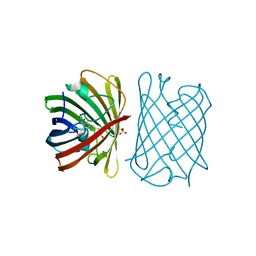 | |
1NFF
 
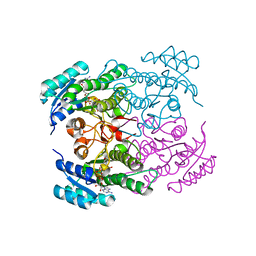 | | Crystal structure of Rv2002 gene product from Mycobacterium tuberculosis | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative oxidoreductase Rv2002 | | Authors: | Yang, J.K, Park, M.S, Waldo, G.S, Suh, S.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-12-14 | | Release date: | 2002-12-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NFR
 
 | | Rv2002 gene product from Mycobacterium tuberculosis | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative oxidoreductase Rv2002 | | Authors: | Yang, J.K, Park, M.S, Waldo, G.S, Suh, S.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-12-16 | | Release date: | 2002-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NFQ
 
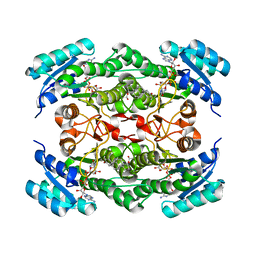 | | Rv2002 gene product from Mycobacterium tuberculosis | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Androsterone, Putative oxidoreductase Rv2002 | | Authors: | Yang, J.K, Park, M.S, Waldo, G.S, Suh, S.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-12-15 | | Release date: | 2002-12-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4M48
 
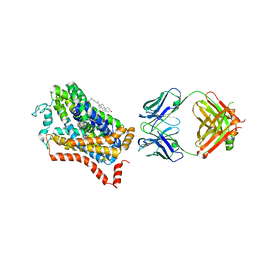 | | X-ray structure of dopamine transporter elucidates antidepressant mechanism | | Descriptor: | 9D5 antibody, heavy chain, light chain, ... | | Authors: | Gouaux, E, Penmatsa, A, Wang, K. | | Deposit date: | 2013-08-06 | | Release date: | 2013-09-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | X-ray structure of dopamine transporter elucidates antidepressant mechanism.
Nature, 503, 2013
|
|
8VDR
 
 | |
8H0L
 
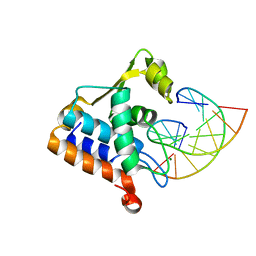 | | Sulfur binding domain of Hga complexed with phosphorothioated DNA | | Descriptor: | DNA (5'-D(*CP*GP*AP*GP*(PST)P*TP*CP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*GP*AP*AP*CP*TP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Liu, G, He, X, Hu, W, Yang, B, Xiao, Q. | | Deposit date: | 2022-09-29 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of a promiscuous DNA sulfur binding domain and application in site-directed RNA base editing.
Nucleic Acids Res., 51, 2023
|
|
8HAS
 
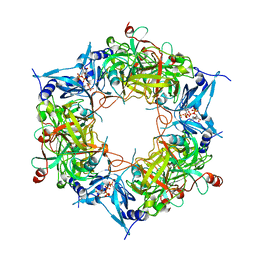 | | NARROW LEAF 1-close from Japonica | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Protein NARROW LEAF 1 | | Authors: | Zhang, S.J, He, Y.J, Wang, N, Zhang, W.J, Liu, C.M. | | Deposit date: | 2022-10-26 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | NARROW LEAF 1-close from Japonica
To Be Published
|
|
