1QZ9
 
 | | The Three Dimensional Structure of Kynureninase from Pseudomonas fluorescens | | Descriptor: | 3,6,9,12,15-PENTAOXAHEPTADECANE, CHLORIDE ION, KYNURENINASE, ... | | Authors: | Momany, C, Levdikov, V, Blagova, L, Lima, S, Phillips, R.S. | | Deposit date: | 2003-09-16 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Three-Dimensional Structure of Kynureninase from Pseudomonas fluorescens.
Biochemistry, 43, 2004
|
|
6FBO
 
 | | Human Methionine Adenosyltransferase II mutant (S114A) in I222 crystal form | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, ADENOSINE, ... | | Authors: | Panmanee, J, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-12-19 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Control and regulation of S-Adenosylmethionine biosynthesis by the regulatory beta subunit and quinolone-based compounds.
Febs J., 286, 2019
|
|
6FCD
 
 | | Human Methionine Adenosyltransferase II mutant (R264A) | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Panmanee, J, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-12-20 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Control and regulation of S-Adenosylmethionine biosynthesis by the regulatory beta subunit and quinolone-based compounds.
Febs J., 286, 2019
|
|
1DD3
 
 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L12 FROM THERMOTOGA MARITIMA | | Descriptor: | 50S RIBOSOMAL PROTEIN L7/L12 | | Authors: | Wahl, M.C, Bourenkov, G.P, Bartunik, H.D, Huber, R. | | Deposit date: | 1999-11-08 | | Release date: | 2000-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flexibility, conformational diversity and two dimerization modes in complexes of ribosomal protein L12.
EMBO J., 19, 2000
|
|
1PH8
 
 | |
1PH9
 
 | |
1PVX
 
 | | DO-1,4-BETA-XYLANASE, ROOM TEMPERATURE, PH 4.5 | | Descriptor: | PROTEIN (ENDO-1,4-BETA-XYLANASE) | | Authors: | Rajeshkumar, P, Eswaramoorthy, S, Vithayathil, P.J, Viswamitra, M.A. | | Deposit date: | 1998-10-20 | | Release date: | 1999-10-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The tertiary structure at 1.59 A resolution and the proposed amino acid sequence of a family-11 xylanase from the thermophilic fungus Paecilomyces varioti bainier.
J.Mol.Biol., 295, 2000
|
|
7B73
 
 | | Insight into the molecular determinants of thermal stability in halohydrin dehalogenase HheD2. | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Short-chain dehydrogenase/reductase SDR | | Authors: | Wessel, J, Petrillo, G, Estevez, M, Bosh, S, Seeger, M, Dijkman, W.P, Hidalgo, A, Uson, I, Osuna, S, Schallmey, A. | | Deposit date: | 2020-12-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into the molecular determinants of thermal stability in halohydrin dehalogenase HheD2.
Febs J., 288, 2021
|
|
5O2F
 
 | | Crystal structure of NDM-1 in complex with hydrolyzed ampicillin - new refinement | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2017-05-20 | | Release date: | 2018-12-26 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5AES
 
 | | Crystal Structure of murine Chronophin (Pyridoxal Phosphate Phosphatase) in Complex with a PNP-derived Inhibitor | | Descriptor: | GLYCEROL, MAGNESIUM ION, PYRIDOXAL PHOSPHATE PHOSPHATASE, ... | | Authors: | Knobloch, G, Jabari, N, Koehn, M, Gohla, A, Schindelin, H. | | Deposit date: | 2015-01-09 | | Release date: | 2015-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Synthesis of Hydrolysis-Resistant Pyridoxal 5'-Phosphate Analogs and Their Biochemical and X-Ray Crystallographic Characterization with the Pyridoxal Phosphatase Chronophin.
Bioorg.Med.Chem., 23, 2015
|
|
5YNZ
 
 | | Crystal structure of the dihydroorotase domain (K1556A) of human CAD | | Descriptor: | CAD protein, ZINC ION | | Authors: | Huang, Y.H, Chen, K.L, Cheng, J.H, Huang, C.Y. | | Deposit date: | 2017-10-26 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.774 Å) | | Cite: | Crystal structures of monometallic dihydropyrimidinase and the human dihydroorotase domain K1556A mutant reveal no lysine carbamylation within the active site
Biochem. Biophys. Res. Commun., 505, 2018
|
|
4TYD
 
 | | Structure-based design of a novel series of azetidine inhibitors of the hepatitis C virus NS3/4A serine protease | | Descriptor: | (4R,6S,7Z,15S,17S)-17-[({7-methoxy-2-[4-(propan-2-yl)-1,3-thiazol-2-yl]quinolin-4-yl}oxy)methyl]-13-methyl-N-[(1-methylcyclopropyl)sulfonyl]-2,14-dioxo-1,3,13-triazatricyclo[13.2.0.0~4,6~]heptadec-7-ene-4-carboxamide, CHLORIDE ION, NS3 protease, ... | | Authors: | Parsy, C. | | Deposit date: | 2014-07-08 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure-based design of a novel series of azetidine inhibitors of the hepatitis C virus NS3/4A serine protease.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3HPA
 
 | | Crystal structure of an amidohydrolase gi:44264246 from an evironmental sample of sargasso sea | | Descriptor: | AMIDOHYDROLASE, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-03 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The hunt for 8-oxoguanine deaminase.
J.Am.Chem.Soc., 132, 2010
|
|
1TQN
 
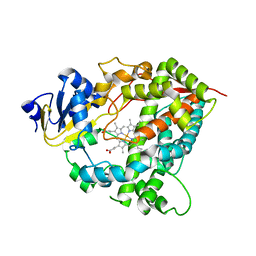 | | Crystal Structure of Human Microsomal P450 3A4 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 3A4 | | Authors: | Yano, J.K, Wester, M.R, Schoch, G.A, Griffin, K.J, Stout, C.D, Johnson, E.F. | | Deposit date: | 2004-06-17 | | Release date: | 2004-07-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Structure of Human Microsomal Cytochrome P450 3A4 Determined by X-ray Crystallography to 2.05-A Resolution
J.Biol.Chem., 279, 2004
|
|
1WD0
 
 | | Crystal structures of the hyperthermophilic chromosomal protein Sac7d in complex with DNA decamers | | Descriptor: | 5'-D(*CP*CP*TP*AP*TP*AP*TP*AP*GP*G)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Ko, T.-P, Chu, H.-M, Chen, C.-Y, Chou, C.-C, Wang, A.H.-J. | | Deposit date: | 2004-05-10 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the hyperthermophilic chromosomal protein Sac7d in complex with DNA decamers.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
12E8
 
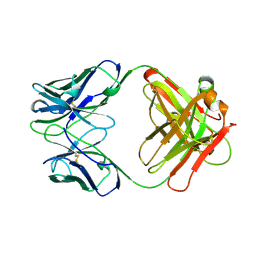 | | 2E8 FAB FRAGMENT | | Descriptor: | IGG1-KAPPA 2E8 FAB (HEAVY CHAIN), IGG1-KAPPA 2E8 FAB (LIGHT CHAIN) | | Authors: | Rupp, B, Trakhanov, S. | | Deposit date: | 1998-03-14 | | Release date: | 1998-08-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a monoclonal 2E8 Fab antibody fragment specific for the low-density lipoprotein-receptor binding region of apolipoprotein E refined at 1.9 A.
Acta Crystallogr.,Sect.D, null, 1999
|
|
1R48
 
 | | Solution structure of the C-terminal cytoplasmic domain residues 468-497 of Escherichia coli protein ProP | | Descriptor: | Proline/betaine transporter | | Authors: | Zoetewey, D.L, Tripet, B.P, Kutateladze, T.G, Overduin, M.J, Wood, J.M, Hodges, R.S. | | Deposit date: | 2003-10-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Antiparallel Coiled-coil Domain from Escherichia coli Osmosensor ProP.
J.Mol.Biol., 334, 2003
|
|
5Z09
 
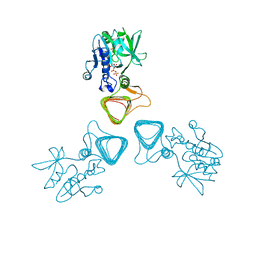 | | ST0452(Y97N)-UTP binding form | | Descriptor: | Dual sugar-1-phosphate nucleotidylyltransferase, URIDINE 5'-TRIPHOSPHATE | | Authors: | Honda, Y, Nakano, S, Ito, S, Dadashipour, M, Zhang, Z, Kawarabayasi, Y. | | Deposit date: | 2017-12-19 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Improvement of ST0452N-Acetylglucosamine-1-Phosphate Uridyltransferase Activity by the Cooperative Effect of Two Single Mutations Identified through Structure-Based Protein Engineering
Appl. Environ. Microbiol., 84, 2018
|
|
1BHC
 
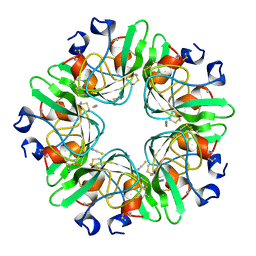 | |
1WD1
 
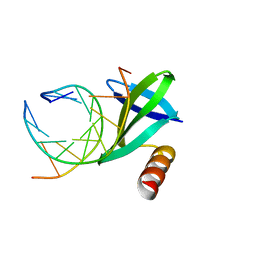 | | Crystal structures of the hyperthermophilic chromosomal protein Sac7d in complex with DNA decamers | | Descriptor: | 5'-D(*CP*CP*TP*AP*CP*GP*TP*AP*GP*G)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Ko, T.-P, Chu, H.-M, Chen, C.-Y, Chou, C.-C, Wang, A.H.-J. | | Deposit date: | 2004-05-10 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the hyperthermophilic chromosomal protein Sac7d in complex with DNA decamers.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PH5
 
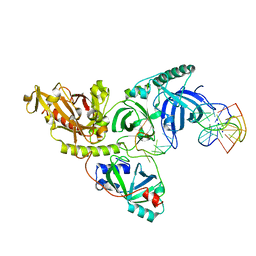 | |
1PH7
 
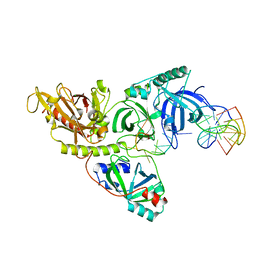 | |
1BDK
 
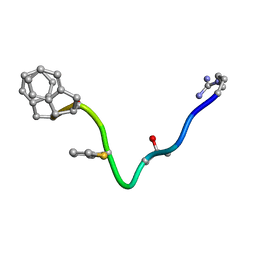 | | AN NMR, CD, MOLECULAR DYNAMICS, AND FLUOROMETRIC STUDY OF THE CONFORMATION OF THE BRADYKININ ANTAGONIST B-9340 IN WATER AND IN AQUEOUS MICELLAR SOLUTIONS | | Descriptor: | bradykinin antagonist B-9340 | | Authors: | Sejbal, J, Kotovych, G, Cann, J.R, Stewart, J.M, Gera, L. | | Deposit date: | 1995-07-28 | | Release date: | 1995-12-07 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | An NMR, CD, molecular dynamics, and fluorometric study of the conformation of the bradykinin antagonist B-9340 in water and in aqueous micellar solutions.
J.Med.Chem., 39, 1996
|
|
7DZ5
 
 | | Crystal structures of D-allulose 3-epimerase with D-sorbose from Sinorhizobium fredii | | Descriptor: | D-sorbose, D-tagatose 3-epimerase, MAGNESIUM ION, ... | | Authors: | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | Deposit date: | 2021-01-23 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substantial Improvement of an Epimerase for the Synthesis of D-Allulose by Biosensor-Based High-Throughput Microdroplet Screening
Angew.Chem.Int.Ed.Engl., 2023
|
|
6KTK
 
 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, complexed with NADH and L-glucono-1,5-lactone, from Paracoccus laeviglucosivorans | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-glucono-1,5-lactone, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity, ... | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
