9C2C
 
 | | Cryo-EM structure of the human P2X1 receptor in the NF449-bound inhibited state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,4',4'',4'''-{carbonylbis[azanediylbenzene-5,1,3-triylbis(carbonylazanediyl)]}tetra(benzene-1,3-disulfonic acid), P2X purinoceptor 1 | | Authors: | Oken, A.C, Lisi, N.E, Ditter, I.A, Shi, H, Mansoor, S.E. | | Deposit date: | 2024-05-30 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of the human P2X1 receptor reveal subtype-specific architecture and antagonism by supramolecular ligand-binding.
Nat Commun, 15, 2024
|
|
5IOF
 
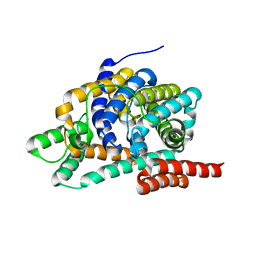 | |
9FWC
 
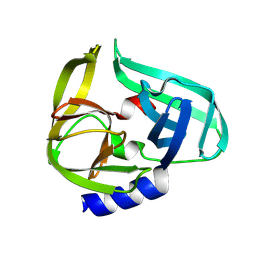 | | Coxsackievirus B3 3C protease in C121 spacegroup | | Descriptor: | Genome polyprotein | | Authors: | Fairhead, M, Lithgo, R.M, MacLean, E.M, Bowesman-Jones, H, Aschenbrenner, J.C, Balcomb, B.H, Capkin, E, Chandran, A.V, Godoy, A.S, Marples, P.G, Fearon, D, von Delft, F, Koekemoer, L. | | Deposit date: | 2024-06-28 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Coxsackievirus B3 3C protease in C121 spacegroup
To Be Published
|
|
9F3D
 
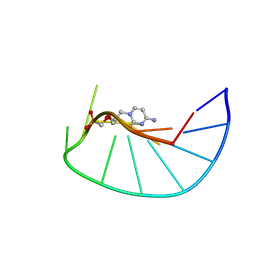 | |
7STK
 
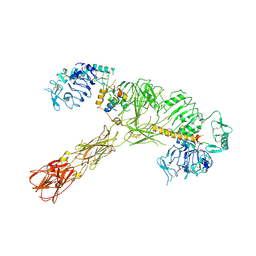 | |
9B9O
 
 | | Crystal structure of FlcD from Pseudomonas aeruginosa bond to iron(II) and substrate | | Descriptor: | (2R)-2-{[(2Z)-2-(hydroxyimino)ethyl]sulfanyl}butanedioic acid, FE (II) ION, Pyrroloquinoline quinone (Coenzyme PQQ) biosynthesis protein C | | Authors: | Walker, M.E, Grove, T.L, Li, B, Redinbo, M.R. | | Deposit date: | 2024-04-02 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Basis for Methine Excision by a Heme Oxygenase-like Enzyme.
Acs Cent.Sci., 10, 2024
|
|
9BJ1
 
 | | Crystal structure of inhibitor GNE-6893 bound to HPK1 | | Descriptor: | (4S,5R,7R,11aP)-10-{[(3R)-3-hydroxy-1-methyl-2-oxopyrrolidin-3-yl]ethynyl}-N~3~-methyl-6,7-dihydro-5H-5,7-methanoimidazo[2,1-a][2]benzazepine-2,3-dicarboxamide, (9S)-2-{[(6P)-8-amino-6-(5-amino-4-methylpyridin-3-yl)-7-fluoroisoquinolin-3-yl]amino}-6-methyl-5,6-dihydro-4H-pyrazolo[1,5-d][1,4]diazepin-7(8H)-one, 1,2-ETHANEDIOL, ... | | Authors: | Kiefer, J.R, Tellis, J.C, Chan, B.K, Wang, W, Wu, P, Choo, E.F, Heffron, T.P, Wei, B, Siu, M. | | Deposit date: | 2024-04-24 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of GNE-6893, a Potent, Selective, Orally Bioavailable Small Molecule Inhibitor of HPK1.
Acs Med.Chem.Lett., 15, 2024
|
|
7STH
 
 | |
9G9S
 
 | | Crystal structure of PbdA bound to veratrate | | Descriptor: | 3,4-dimethoxybenzoic acid, Cytochrome P450 CYP199, GLYCEROL, ... | | Authors: | Hinchen, D.J, Wolf, M.E, Eltis, L.D, McGeehan, J.E. | | Deposit date: | 2024-07-25 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterization of a cytochrome P450 that catalyzes the O-demethylation of lignin-derived benzoates.
J.Biol.Chem., 2024
|
|
9EUU
 
 | | Structure of recombinant alpha-synuclein fibrils 1B capable of seeding GCIs in vivo | | Descriptor: | Alpha-synuclein | | Authors: | Burger, D, Kashyrina, M, Lewis, A, De Nuccio, F, Mohammed, I, de La Seigliere, H, van den Heuvel, L, Feuillie, C, Verchere, J, Berbon, M, Arotcarena, M, Retailleau, A, Bezard, E, Laferriere, F, Loquet, A, Bousset, L, Baron, T, Lofrumento, D.D, De Giorgi, F, Stahlberg, H, Ichas, F. | | Deposit date: | 2024-03-28 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | Multiple System Atrophy: Insights from aSyn Fibril Structure
To Be Published
|
|
6GQW
 
 | | KRAS-169 Q61H GPPNHP + CH-1 | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Cruz-Migoni, A, Quevedo, C.E, Carr, S.B, Phillips, S.E.V, Rabbitts, T.H. | | Deposit date: | 2018-06-08 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6QS1
 
 | |
8VIW
 
 | | Cryo-EM structure of heparosan synthase 2 from Pasteurella multocida with polysaccharide in the GlcNAc-T active site | | Descriptor: | Heparosan synthase B, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Krahn, J.M, Pedersen, L.C, Liu, J, Stancanelli, E, Borgnia, M, Vivarette, E. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and Functional Analysis of Heparosan Synthase 2 from Pasteurella multocida to Improve the Synthesis of Heparin
Acs Catalysis, 14, 2024
|
|
5IK4
 
 | | Laminin A2LG45 C-form, Apo. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Briggs, D.C, Hohenester, E, Campbell, K.P. | | Deposit date: | 2016-03-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural basis of laminin binding to the LARGE glycans on dystroglycan.
Nat.Chem.Biol., 12, 2016
|
|
6VUC
 
 | |
9AT8
 
 | | Fab 77-stabilized MeV F ectodomain fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, mAB 77 heavy chain, ... | | Authors: | Zyla, D, Saphire, E.O. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A neutralizing antibody prevents postfusion transition of measles virus fusion protein.
Science, 384, 2024
|
|
8VSX
 
 | | NMR Structure of GCAP5 R22A | | Descriptor: | Guanylyl cyclase-activating protein 1 | | Authors: | Cudia, D.L, Ames, J.B. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Retinal Guanylate Cyclase Activating Protein 5 (GCAP5) with R22A Mutation That Abolishes Dimerization and Enhances Cyclase Activation.
Biochemistry, 63, 2024
|
|
8ZMY
 
 | | F0502B-bound WT polymorph 5a alpha-synuclein fibril | | Descriptor: | 2-bromanyl-4-[(~{E})-2-[6-[2-(2-fluoranylethoxy)ethyl-methyl-amino]-5-methyl-1,3-benzothiazol-2-yl]ethenyl]phenol, Alpha-synuclein | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2024-05-24 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9B9N
 
 | | Crystal structure of FlcD from Pseudomonas aeruginosa bound to iron (II) and substrate | | Descriptor: | (2R)-2-{[(2Z)-2-(hydroxyimino)ethyl]sulfanyl}butanedioic acid, FE (III) ION, Pyrroloquinoline quinone (Coenzyme PQQ) biosynthesis protein C | | Authors: | Walker, M.E, Grove, T.L, Li, B, Redinbo, M.R. | | Deposit date: | 2024-04-02 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Basis for Methine Excision by a Heme Oxygenase-like Enzyme.
Acs Cent.Sci., 10, 2024
|
|
9G9R
 
 | | Crystal structure of PbdA bound to p-ethylbenzoate | | Descriptor: | 4-ethylbenzoic acid, Cytochrome P450 CYP199, GLYCEROL, ... | | Authors: | Hinchen, D.J, Wolf, M.E, Eltis, L.D, McGeehan, J.E. | | Deposit date: | 2024-07-25 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Characterization of a cytochrome P450 that catalyzes the O-demethylation of lignin-derived benzoates.
J.Biol.Chem., 2024
|
|
7PHZ
 
 | | Crystal structure of X77 bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup P2(1)2(1)2(1). | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8W04
 
 | |
7PKA
 
 | | Synechocystis sp. PCC6803 glutathione transferase Chi 1, GSOH bound | | Descriptor: | Glutathione S-transferase, POTASSIUM ION, S-Hydroxy-Glutathione | | Authors: | Didierjean, C, Mocchetti, E, Hecker, A, Favier, F. | | Deposit date: | 2021-08-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure and functions of glutathione transferase Chi 1 from cyanobacterium Synechocystis sp. PCC 6803
To Be Published
|
|
6QPF
 
 | | Influenza A virus Polymerase Heterotrimer A/duck/Fujian/01/2002(H5N1) | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Fan, H.T, Keown, J.R, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-02-13 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.634 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
9FYD
 
 | | tubulin - cryptophycin-uD[Dab] complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Dessin, C, Schachtsiek, T, Voss, J, Abel, A.-C, Neumann, B, Stammler, H.-G, Prota, A.E, Sewald, N. | | Deposit date: | 2024-07-03 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Highly Cytotoxic Cryptophycin Derivatives with Modification in Unit D for Conjugation.
Angew.Chem.Int.Ed.Engl., 2024
|
|
