8F0N
 
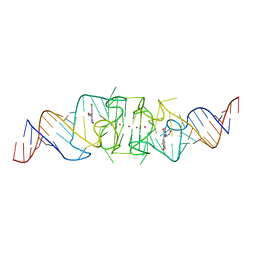 | | Wobble Beetroot (A16U-U38G) dimer bound to DFHO | | Descriptor: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, POTASSIUM ION, RNA (49-MER) | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-11-03 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Co-crystal structures of the fluorogenic aptamer Beetroot show that close homology may not predict similar RNA architecture.
Nat Commun, 14, 2023
|
|
2AR3
 
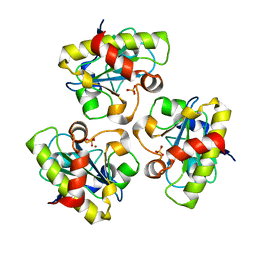 | | E90A mutant structure of PlyL | | Descriptor: | PHOSPHATE ION, ZINC ION, prophage lambdaba02, ... | | Authors: | Low, L.Y, Yang, C, Perego, M, Osterman, A, Liddington, R.C. | | Deposit date: | 2005-08-19 | | Release date: | 2006-06-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and lytic activity of a Bacillus anthracis prophage endolysin.
J.Biol.Chem., 280, 2005
|
|
2POX
 
 | |
2UXV
 
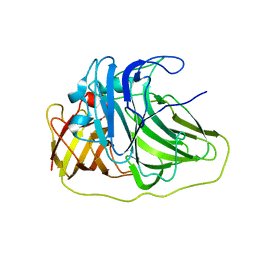 | | SufI Protein from Escherichia Coli | | Descriptor: | PROTEIN SUFI | | Authors: | Tarry, M.J, Roversi, P, Sargent, F, Berks, B.C, Lea, S.M. | | Deposit date: | 2007-03-30 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The Escherichia Coli Cell Division Protein and Model Tat Substrate Sufi (Ftsp) Localizes to the Septal Ring and Has a Multicopper Oxidase-Like Structure.
J.Mol.Biol., 386, 2009
|
|
3E5V
 
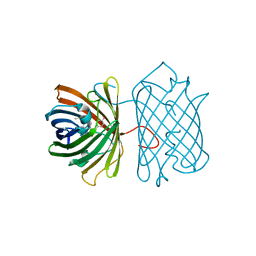 | | Crystal Structure Analysis of eqFP611 Double Mutant T122R, N143S | | Descriptor: | Red fluorescent protein eqFP611 | | Authors: | Nar, H, Nienhaus, K, Nienhaus, U, Wiedenmann, J. | | Deposit date: | 2008-08-14 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Trans-cis isomerization is responsible for the red-shifted fluorescence in variants of the red fluorescent protein eqFP611.
J.Am.Chem.Soc., 130, 2008
|
|
3E5T
 
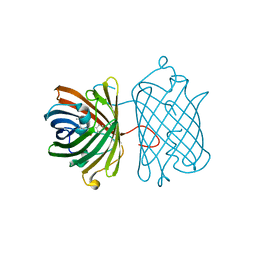 | | Crystal Structure Analysis of FP611 | | Descriptor: | Red fluorescent protein eqFP611 | | Authors: | Nar, H, Nienhaus, K, Nienhaus, U, Wiedenmann, J. | | Deposit date: | 2008-08-14 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Trans-cis isomerization is responsible for the red-shifted fluorescence in variants of the red fluorescent protein eqFP611.
J.Am.Chem.Soc., 130, 2008
|
|
3E5W
 
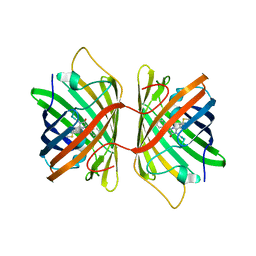 | | Crystal Structure Analysis of FP611 | | Descriptor: | Red fluorescent protein eqFP611 | | Authors: | Nienhaus, K, Nar, H, Heilker, R, Wiedenmann, J, Nienhaus, G.U. | | Deposit date: | 2008-08-14 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Trans-cis isomerization is responsible for the red-shifted fluorescence in variants of the red fluorescent protein eqFP611.
J.Am.Chem.Soc., 130, 2008
|
|
6RNH
 
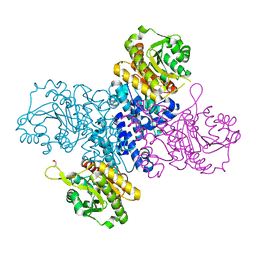 | | Structure of C-terminal truncated Plasmodium falciparum IMP-nucleotidase | | Descriptor: | GLYCEROL, IMP-specific 5'-nucleotidase, putative | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-08 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
6RN1
 
 | | Structure of N-terminal truncated Plasmodium falciparum IMP-nucleotidase | | Descriptor: | IMP-specific 5'-nucleotidase, putative | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
6RMW
 
 | | Structure of N-terminal truncated IMP bound Plasmodium falciparum IMP-nucleotidase | | Descriptor: | GLYCEROL, IMP-specific 5'-nucleotidase, putative, ... | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
6RMO
 
 | | Structure of Plasmodium falciparum IMP-nucleotidase | | Descriptor: | IMP-specific 5'-nucleotidase, putative | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
6RME
 
 | | Structure of IMP bound Plasmodium falciparum IMP-nucleotidase mutant D172N | | Descriptor: | GLYCEROL, IMP-specific 5'-nucleotidase, putative, ... | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-06 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
6RQR
 
 | | Extended NHERF1 PDZ2 domain in complex with the PDZ-binding motif of CFTR | | Descriptor: | Na(+)/H(+) exchange regulatory cofactor NHE-RF1,Cystic fibrosis transmembrane conductance regulator | | Authors: | Martin, E.R, Ford, R.C, Robinson, R.C. | | Deposit date: | 2019-05-16 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In vivocrystals reveal critical features of the interaction between cystic fibrosis transmembrane conductance regulator (CFTR) and the PDZ2 domain of Na+/H+exchange cofactor NHERF1.
J.Biol.Chem., 295, 2020
|
|
6RMD
 
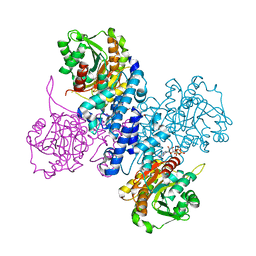 | | Structure of ATP bound Plasmodium falciparum IMP-nucleotidase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, IMP-specific 5'-nucleotidase, ... | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
3U0N
 
 | |
3U0M
 
 | |
3U0L
 
 | | Crystal structure of the engineered fluorescent protein mRuby, crystal form 1, pH 4.5 | | Descriptor: | ACETATE ION, mRuby | | Authors: | Akerboom, J, Looger, L.L, Schreiter, E.R. | | Deposit date: | 2011-09-28 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics.
Front Mol Neurosci, 6, 2013
|
|
8SV0
 
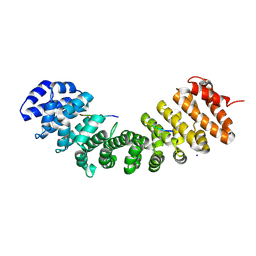 | | The crystal structure of the classical binding interface of Importin alpha 2 and nuclear localisation signal sequence in Psittacine siadenovirus core protein VII | | Descriptor: | Importin subunit alpha-1, SODIUM ION, protein VII | | Authors: | Athukorala, A, Sarker, S, Forwood, J.K, Donnelly, C.M. | | Deposit date: | 2023-05-14 | | Release date: | 2023-05-31 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional characterization of siadenovirus core protein VII nuclear localization demonstrates the existence of multiple nuclear transport pathways.
J.Gen.Virol., 105, 2024
|
|
2LZP
 
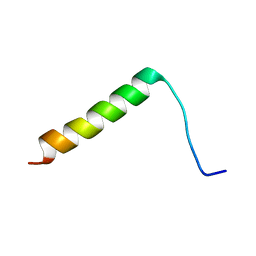 | |
8TGP
 
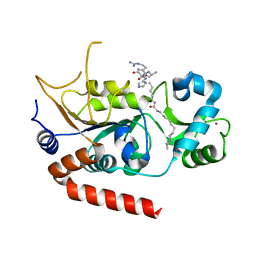 | |
8TU9
 
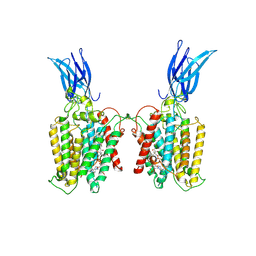 | | Cryo-EM structure of HGSNAT-acetyl-CoA complex at pH 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, Enhanced green fluorescent protein,Heparan-alpha-glucosaminide N-acetyltransferase,Isoform 2 of Heparan-alpha-glucosaminide N-acetyltransferase | | Authors: | Navratna, V, Kumar, A, Mosalaganti, S. | | Deposit date: | 2023-08-15 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure of the human heparan-alpha-glucosaminide N -acetyltransferase (HGSNAT).
Elife, 13, 2024
|
|
4IR4
 
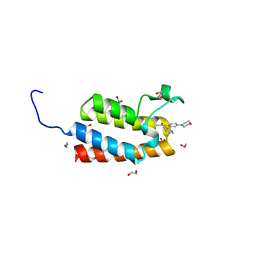 | | Crystal Structure of the bromodomain of human BAZ2B in complex with 1-[7-(morpholin-4-yl)-1-(pyridin-2-yl)indolizin-3-yl]ethanone (GSK2834113A) | | Descriptor: | 1,2-ETHANEDIOL, 1-[7-(morpholin-4-yl)-1-(pyridin-2-yl)indolizin-3-yl]ethanone, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, Chung, C.W, Drewry, D, Chen, P, Filippakopoulos, P, Fedorov, O, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery and Characterization of GSK2801, a Selective Chemical Probe for the Bromodomains BAZ2A and BAZ2B.
J.Med.Chem., 59, 2016
|
|
2MKB
 
 | |
4IR5
 
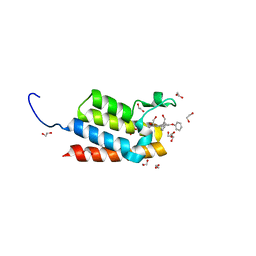 | | Crystal Structure of the bromodomain of human BAZ2B in complex with 1-{1-[2-(hydroxymethyl)phenyl]-7-phenoxyindolizin-3-yl}ethanone (GSK2847449A) | | Descriptor: | 1,2-ETHANEDIOL, 1-{1-[2-(hydroxymethyl)phenyl]-7-phenoxyindolizin-3-yl}ethanone, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, Chung, C.W, Drewry, D, Chen, P, Filippakopoulos, P, Fedorov, O, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and Characterization of GSK2801, a Selective Chemical Probe for the Bromodomains BAZ2A and BAZ2B.
J.Med.Chem., 59, 2016
|
|
4IR3
 
 | | Crystal Structure of the bromodomain of human BAZ2B in complex with 1-[7-amino-1-(pyrimidin-2-yl)indolizin-3-yl]ethanone (GSK2833282A) | | Descriptor: | 1,2-ETHANEDIOL, 1-[7-amino-1-(pyrimidin-2-yl)indolizin-3-yl]ethanone, Bromodomain adjacent to zinc finger domain protein 2B, ... | | Authors: | Chaikuad, A, Felletar, I, Chung, C.W, Drewry, D, Chen, P, Filippakopoulos, P, Fedorov, O, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and Characterization of GSK2801, a Selective Chemical Probe for the Bromodomains BAZ2A and BAZ2B.
J.Med.Chem., 59, 2016
|
|
