1UK3
 
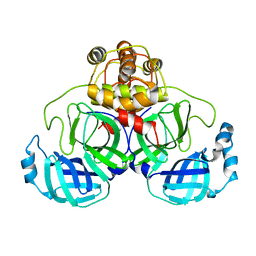 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) At pH7.6 | | Descriptor: | 3C-like proteinase | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-08-14 | | Release date: | 2003-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
7VLO
 
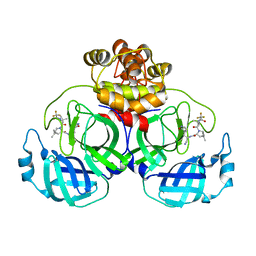 | | Crystal structure of SARS coronavirus main protease in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2021-10-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.0227 Å) | | Cite: | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VLP
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07321332 in spacegroup P1211 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, Replicase polyprotein 1a | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Li, J, Zhang, J. | | Deposit date: | 2021-10-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.50251937 Å) | | Cite: | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
1UK4
 
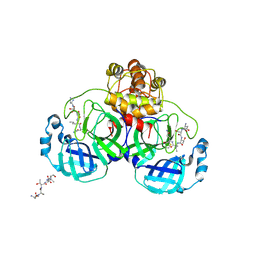 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) Complexed With An Inhibitor | | Descriptor: | 3C-like proteinase nsp5, 5-mer peptide of inhibitor | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-08-14 | | Release date: | 2003-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
3C0L
 
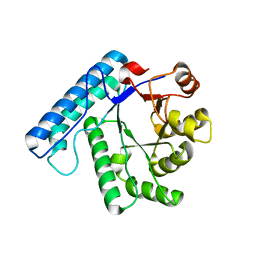 | | UVDE K229R | | Descriptor: | UV endonuclease | | Authors: | Meulenbroek, E.M, Paspaleva, K, Thomassen, E.A.J, Abrahams, J.P, Goosen, N, Pannu, N.S. | | Deposit date: | 2008-01-21 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Involvement of a carboxylated lysine in UV damage endonuclease
Protein Sci., 18, 2009
|
|
3C0S
 
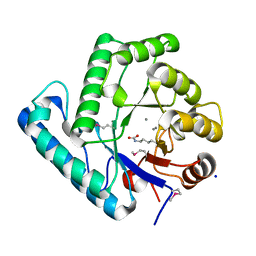 | | UVDE 3 metals | | Descriptor: | MANGANESE (II) ION, SODIUM ION, UV endonuclease | | Authors: | Meulenbroek, E.M, Paspaleva, K, Thomassen, E.A.J, Abrahams, J.P, Goosen, N, Pannu, N.S. | | Deposit date: | 2008-01-21 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Involvement of a carboxylated lysine in UV damage endonuclease
Protein Sci., 18, 2009
|
|
7W7F
 
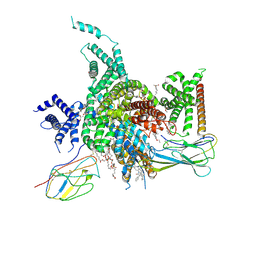 | | Cryo-EM structure of human NaV1.3/beta1/beta2-ICA121431 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2,2-diphenyl-~{N}-[4-(1,3-thiazol-2-ylsulfamoyl)phenyl]ethanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D, Li, X. | | Deposit date: | 2021-12-04 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis for modulation of human Na V 1.3 by clinical drug and selective antagonist.
Nat Commun, 13, 2022
|
|
7W77
 
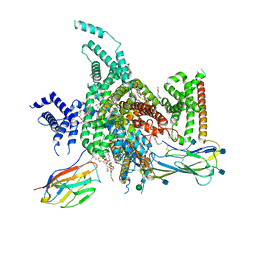 | | cryo-EM structure of human NaV1.3/beta1/beta2-bulleyaconitineA | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D, Li, X. | | Deposit date: | 2021-12-03 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for modulation of human Na V 1.3 by clinical drug and selective antagonist.
Nat Commun, 13, 2022
|
|
2V6A
 
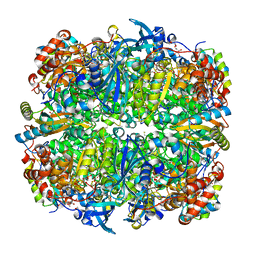 | | Crystal structure of Chlamydomonas reinhardtii Rubisco with large- subunit mutations V331A, G344S | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Karkehabadi, S, Satagopan, S, Taylor, T.C, Spreitzer, R.J, Andersson, I. | | Deposit date: | 2007-07-14 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of Altered Large-Subunit Loop-6-Carboxy-Terminus Interactions that Influence Catalytic Efficiency and Co2-O2 Specificity of Ribulose-1,5-Bisphosphate Carboxylase Oxygenase
Biochemistry, 46, 2007
|
|
5ESE
 
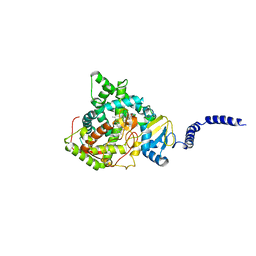 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) G73R mutant complexed with fluconazole | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sagatova, A, Keniya, M.V, Wilson, R.K, Sabherwal, M, Tyndall, J.D.A, Monk, B.C. | | Deposit date: | 2015-11-16 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Impact of Homologous Resistance Mutations from Pathogenic Yeast on Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase.
Antimicrob.Agents Chemother., 62, 2018
|
|
1D44
 
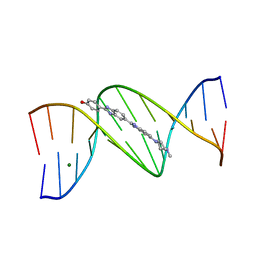 | | DNA DODECAMER C-G-C-G-A-A-T-T-C-G-C-G/HOECHST 33258 COMPLEX: 0 DEGREES C, PIPERAZINE DOWN | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Lipanov, A.A, Dickerson, R.E. | | Deposit date: | 1991-06-04 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G.
Biochemistry, 30, 1991
|
|
2VDI
 
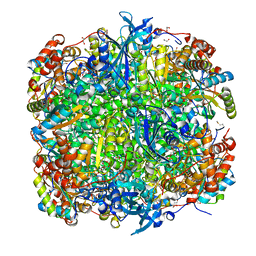 | | Crystal structure of Chlamydomonas reinhardtii Rubisco with a large- subunit C192S mutation | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Garcia-Murria, M.-J, Karkehabadi, S, Marin-Navarro, J, Satagopan, S, Andersson, I, Spreitzer, R.J, Moreno, J. | | Deposit date: | 2007-10-09 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and Functional Consequences of the Replacement of Proximal Residues Cys-172 and Cys-192 in the Large Subunit of Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase from Chlamydomonas Reinhardtii
Biochem.J., 411, 2008
|
|
2W51
 
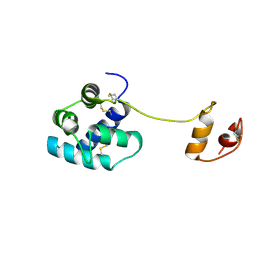 | | Human mesencephalic astrocyte-derived neurotrophic factor (MANF) | | Descriptor: | PROTEIN ARMET | | Authors: | Parkash, V, Lindholm, P, Peranen, J, Kalkkinen, N, Oksanen, E, Saarma, M, Leppanen, V.M, Goldman, A. | | Deposit date: | 2008-12-03 | | Release date: | 2009-03-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of the Conserved Neurotrophic Factors Manf and Cdnf Explains Why They are Bifunctional.
Protein Eng.Des.Sel., 22, 2009
|
|
2VXH
 
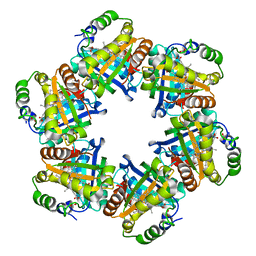 | | The crystal structure of chlorite dismutase: a detox enzyme producing molecular oxygen | | Descriptor: | CARBONATE ION, CHLORITE DISMUTASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | De Geus, D.C, Thomassen, E.A.J, Hagedoorn, P.L, Pannu, N.S, Abrahams, J.P. | | Deposit date: | 2008-07-04 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Chlorite Dismutase, a Detoxifying Enzyme Producing Molecular Oxygen
J.Mol.Biol., 387, 2009
|
|
2WHR
 
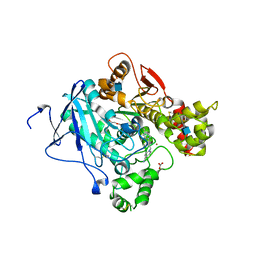 | | Crystal structure of acetylcholinesterase in complex with K027 | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Ekstrom, F, Hornberg, A, Artursson, E, Hammarstrom, L.-G, Schneider, G, Pang, Y.-P. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Structure of Hi-6Sarin-Acetylcholinesterase Determined by X-Ray Crystallography and Molecular Dynamics Simulation: Reactivator Mechanism and Design.
Plos One, 4, 2009
|
|
4X3B
 
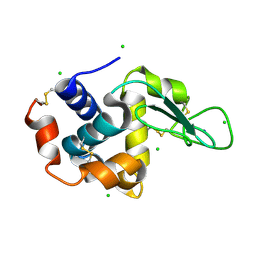 | | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Roedig, P, Vartiainen, I, Duman, R, Panneerselvam, S, Stuebe, N, Lorbeer, O, Warmer, M, Sutton, G, Stuart, D.I, Weckert, E, David, C, Wagner, A, Meents, A. | | Deposit date: | 2014-11-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering.
Sci Rep, 5, 2015
|
|
4OIU
 
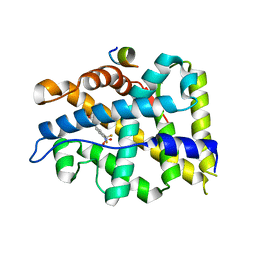 | | Crystal structure of T877A-AR-LBD bound with co-regulator peptide | | Descriptor: | Androgen receptor, HYDROXYFLUTAMIDE, co-regulator peptide | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-20 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
1VBN
 
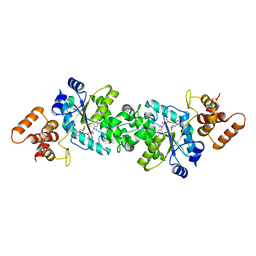 | | Escherichia coli tyrosyl-tRNA synthetase mutant complexed with Tyr-AMS | | Descriptor: | 5'-O-[N-(L-TYROSYL)SULFAMOYL]ADENOSINE, Tyrosyl-tRNA synthetase | | Authors: | Kobayashi, T, Sakamoto, K, Takimura, T, Kamata, K, Sekine, R, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-27 | | Release date: | 2005-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of nonnatural amino acid recognition by an engineered aminoacyl-tRNA synthetase for genetic code expansion
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1UZ8
 
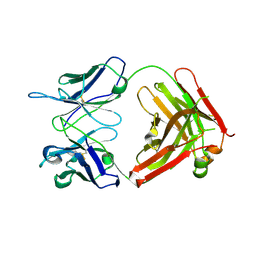 | | anti-Lewis X Fab fragment in complex with Lewis X | | Descriptor: | IGG FAB (IGG3, KAPPA) HEAVY CHAIN 291-2G3-A, KAPPA) LIGHT CHAIN 291-2G3-A, ... | | Authors: | Van Roon, A.M.M, Pannu, N.S, De Vrind, J.P.M, Hokke, C.H, Deelder, A.M, Van Der marel, G.A, Van Boom, J.H, Abrahams, J.P. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of an Anti-Lewis X Fab Fragment in Complex with its Lewis X Antigen
Structure, 12, 2004
|
|
1D45
 
 | | DNA DODECAMER C-G-C-G-A-A-T-T-C-G-C-G/HOECHST 33258 COMPLEX:-25 DEGREES C, PIPERAZINE DOWN | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Lipanov, A.A, Dickerson, R.E. | | Deposit date: | 1991-06-04 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G.
Biochemistry, 30, 1991
|
|
1CKR
 
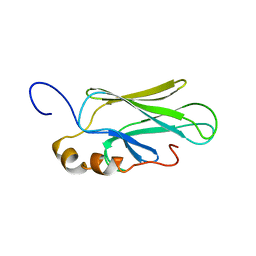 | | HIGH RESOLUTION SOLUTION STRUCTURE OF THE HEAT SHOCK COGNATE-70 KD SUBSTRATE BINDING DOMAIN OBTAINED BY MULTIDIMENSIONAL NMR TECHNIQUES | | Descriptor: | HEAT SHOCK SUBSTRATE BINDING DOMAIN OF HSC-70 | | Authors: | Morshauser, R.C, Hu, W, Wang, H, Pang, Y, Flynn, G.C, Zuiderweg, E.R.P. | | Deposit date: | 1999-04-22 | | Release date: | 1999-04-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of the 18 kDa substrate-binding domain of the mammalian chaperone protein Hsc70.
J.Mol.Biol., 289, 1999
|
|
4X35
 
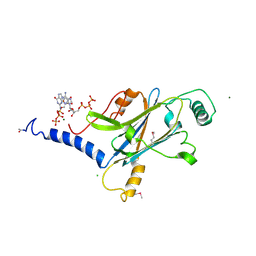 | | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Roedig, P, Vartiainen, I, Duman, R, Panneerselvam, S, Stuebe, N, Lorbeer, O, Warmer, M, Sutton, G, Stuart, D.I, Weckert, E, David, C, Wagner, A, Meents, A. | | Deposit date: | 2014-11-27 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering.
Sci Rep, 5, 2015
|
|
4O34
 
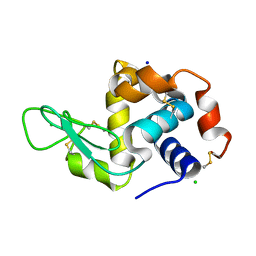 | | Room temperature macromolecular serial crystallography using synchrotron radiation | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Stellato, F, Oberthuer, D, Liang, M, Bean, R, Gati, C, Yefanov, O, Barty, A, Burkhardt, A, Fischer, P, Galli, L, Kirian, R.A, Mayer, J, Pannerselvam, S, Yoon, C.H, Chervinskii, F, Speller, E, White, T.A, Betzel, C, Meents, A, Chapman, H.N. | | Deposit date: | 2013-12-18 | | Release date: | 2014-06-11 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Room-temperature macromolecular serial crystallography using synchrotron radiation.
IUCrJ, 1, 2014
|
|
2Z4E
 
 | |
2WOO
 
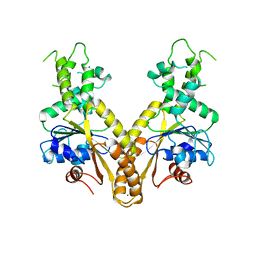 | | Nucleotide-free form of S. pombe Get3 | | Descriptor: | ATPASE GET3, ZINC ION | | Authors: | Mateja, A, Szlachcic, A, Downing, M.E, Dobosz, M, Mariappan, M, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2009-07-27 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.006 Å) | | Cite: | The Structural Basis of Tail-Anchored Membrane Protein Recognition by Get3.
Nature, 461, 2009
|
|
