4J9H
 
 | |
6SDM
 
 | | NADH-dependent variant of TBADH | | Descriptor: | NADP-dependent isopropanol dehydrogenase, ZINC ION | | Authors: | Selles Vidal, L, Murray, J.W, Heap, J.T. | | Deposit date: | 2019-07-28 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Versatile selective evolutionary pressure using synthetic defect in universal metabolism.
Nat Commun, 12, 2021
|
|
6HEQ
 
 | | Prion nanobody 484 | | Descriptor: | Prion nanobody 484 | | Authors: | Soror, S.H, Abskharon, R.N, Wohlkonig, A. | | Deposit date: | 2018-08-20 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structural evidence for the critical role of the prion protein hydrophobic region in forming an infectious prion.
Plos Pathog., 15, 2019
|
|
6SCW
 
 | |
6HF2
 
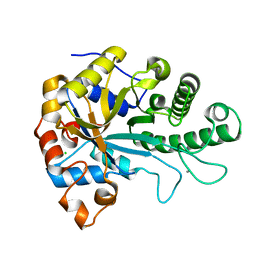 | | The structure of BoMan26B, a GH26 beta-mannanase from Bacteroides ovatus | | Descriptor: | CALCIUM ION, CHLORIDE ION, Glycosyl hydrolase family 26 | | Authors: | Bagenholm, V, Logan, D.T, Stalbrand, H. | | Deposit date: | 2018-08-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A surface-exposed GH26 beta-mannanase fromBacteroides ovatus: Structure, role, and phylogenetic analysis ofBoMan26B.
J.Biol.Chem., 294, 2019
|
|
6HF4
 
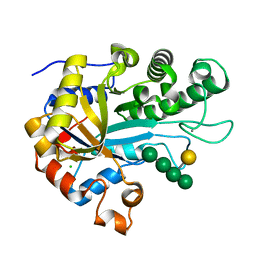 | | The structure of BoMan26B, a GH26 beta-mannanase from Bacteroides ovatus, complexed with G1M4 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Glycosyl hydrolase family 26, ... | | Authors: | Bagenholm, V, Logan, D.T, Stalbrand, H. | | Deposit date: | 2018-08-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | A surface-exposed GH26 beta-mannanase fromBacteroides ovatus: Structure, role, and phylogenetic analysis ofBoMan26B.
J.Biol.Chem., 294, 2019
|
|
8S84
 
 | |
6IPX
 
 | |
7QDU
 
 | |
7QDL
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 with I-BET567 | | Descriptor: | (2S,4R)-1-acetyl-4-((5-chloropyrimidin-2-yl)amino)-2-methyl-1,2,3,4-tetrahydroquinoline-6-carboxamide, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Chung, C. | | Deposit date: | 2021-11-27 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Design, Synthesis, and Characterization of I-BET567, a Pan-Bromodomain and Extra Terminal (BET) Bromodomain Oral Candidate.
J.Med.Chem., 65, 2022
|
|
6IS8
 
 | | Crystal structure of ZmMoc1 D115N mutant in complex with Holliday junction | | Descriptor: | DNA (33-MER), MAGNESIUM ION, Monokaryotic chloroplast 1, ... | | Authors: | Lin, Z, Lin, H, Zhang, D, Yuan, C. | | Deposit date: | 2018-11-15 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis of sequence-specific Holliday junction cleavage by MOC1.
Nat.Chem.Biol., 15, 2019
|
|
8S7H
 
 | | Fructose 6-phosphate aldolase (FSA) from Escherichia coli | | Descriptor: | Fructose-6-phosphate aldolase 1 | | Authors: | Hebert, H, Widersten, M. | | Deposit date: | 2024-03-01 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The Structure of an Engineered Aldolase Catalyzing Carboligation of Arylated Ketones and Aldehydes, Capturing the Iminium Reaction Intermediate, Points Towards a Conserved Tyrosine Residue as Catalytic Acid/Base
To Be Published
|
|
8S7I
 
 | | Fructose 6-phosphate aldolase, L107C/A129G/R134V/L163C/S166G mutant | | Descriptor: | Fructose-6-phosphate aldolase 1 | | Authors: | Hebert, H, Widersten, M. | | Deposit date: | 2024-03-01 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The Structure of an Engineered Aldolase Catalyzing Carboligation of Arylated Ketones and Aldehydes, Capturing the Iminium Reaction Intermediate, Points Towards a Conserved Tyrosine Residue as Catalytic Acid/Base
To Be Published
|
|
5OQ7
 
 | | Structure of CHK1 8-pt. mutant complex with arylbenzamide LRRK2 inhibitor | | Descriptor: | 5-(4-methylpiperazin-1-yl)-2-phenylmethoxy-~{N}-pyridin-3-yl-benzamide, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-10 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
6JKC
 
 | | Crystal structure of tetrameric PepTSo2 in P4212 space group | | Descriptor: | Proton:oligopeptide symporter POT family | | Authors: | Nagamura, R, Fukuda, M, Ishitani, R, Nureki, O. | | Deposit date: | 2019-02-28 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for oligomerization of the prokaryotic peptide transporter PepTSo2.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6JRF
 
 | | Crystal structure of ZmMoc1-Holliday junction Complex in the presence of Calcium | | Descriptor: | CALCIUM ION, DNA (33-MER), Monokaryotic chloroplast 1, ... | | Authors: | Lin, Z, Lin, H, Zhang, D, Yuan, C. | | Deposit date: | 2019-04-03 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Structural basis of sequence-specific Holliday junction cleavage by MOC1.
Nat.Chem.Biol., 15, 2019
|
|
8R4I
 
 | |
1XC6
 
 | | Native Structure Of Beta-Galactosidase from Penicillium sp. in complex with Galactose | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rojas, A.L, Nagem, R.A.P, Neustroev, K.N, Arand, M, Adamska, M, Eneyskaya, E.V, Kulminskaya, A.A, Garratt, R.C, Golubev, A.M, Polikarpov, I. | | Deposit date: | 2004-09-01 | | Release date: | 2004-11-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of beta-Galactosidase from Penicillium sp. and its Complex with Galactose
J.Mol.Biol., 343, 2004
|
|
6RS9
 
 | | X-ray crystal structure of LsAA9B (xylotetraose soak) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AA9, BICINE, ... | | Authors: | Frandsen, K.E.H, Tovborg, M, Poulsen, J.C.N, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2019-05-21 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insights into an unusual Auxiliary Activity 9 family member lacking the histidine brace motif of lytic polysaccharide monooxygenases.
J.Biol.Chem., 294, 2019
|
|
8PON
 
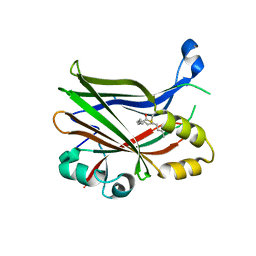 | | TEAD2 in complex with an inhibitor | | Descriptor: | 4-fluoranyl-2-[(3-phenylmethoxyphenyl)amino]benzoic acid, MYRISTIC ACID, Transcriptional enhancer factor TEF-4 | | Authors: | Guichou, J.F. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of LM-41 and AF-2112, two flufenamic acid-derived TEAD inhibitors obtained through the replacement of the trifluoromethyl group by aryl rings.
Bioorg.Med.Chem.Lett., 95, 2023
|
|
8POM
 
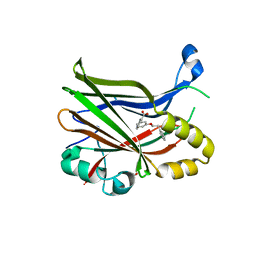 | | TEAD2 in complex with an inhibitor | | Descriptor: | 2-[[3-(2-phenylethoxy)phenyl]amino]pyridine-3-carboxylic acid, GLYCEROL, Transcriptional enhancer factor TEF-4 | | Authors: | Guichou, J.F. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development of LM-41 and AF-2112, two flufenamic acid-derived TEAD inhibitors obtained through the replacement of the trifluoromethyl group by aryl rings.
Bioorg.Med.Chem.Lett., 95, 2023
|
|
8POJ
 
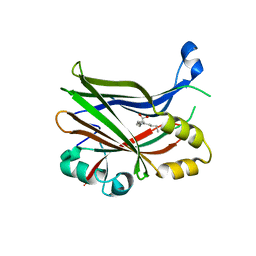 | | TEAD2 in complex with an inhibitor | | Descriptor: | 4-fluoranyl-2-[(3-phenylphenyl)amino]benzoic acid, MYRISTIC ACID, Transcriptional enhancer factor TEF-4 | | Authors: | Guichou, J.F. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Development of LM-41 and AF-2112, two flufenamic acid-derived TEAD inhibitors obtained through the replacement of the trifluoromethyl group by aryl rings.
Bioorg.Med.Chem.Lett., 95, 2023
|
|
5O40
 
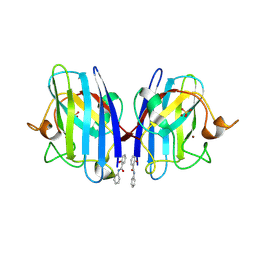 | | SOD1 bound to Ebselen | | Descriptor: | N-phenyl-2-selanylbenzamide, SULFATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Capper, M.J, Wright, G.S.A, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-05-25 | | Release date: | 2018-06-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The cysteine-reactive small molecule ebselen facilitates effective SOD1 maturation.
Nat Commun, 9, 2018
|
|
8TH1
 
 | |
1I10
 
 | | HUMAN MUSCLE L-LACTATE DEHYDROGENASE M CHAIN, TERNARY COMPLEX WITH NADH AND OXAMATE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ACETATE ION, L-LACTATE DEHYDROGENASE M CHAIN, ... | | Authors: | Read, J.A, Winter, V.J, Eszes, C.M, Sessions, R.B, Brady, R.L. | | Deposit date: | 2001-01-30 | | Release date: | 2001-03-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for altered activity of M- and H-isozyme forms of human lactate dehydrogenase.
Proteins, 43, 2001
|
|
