3EQW
 
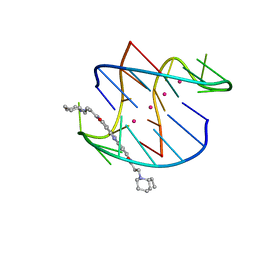 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6042 in small unit cell | | Descriptor: | 3,6-Bis{3-[(2R)-(2-ethylpiperidino)]propionamido}acridine, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', POTASSIUM ION | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
3ET8
 
 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6054 | | Descriptor: | 3,6-Bis{3-(3-[(3R)-methylpiperidino)]propionamido}acridine, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', POTASSIUM ION | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-10-07 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
3E9V
 
 | | Crystal structure of human B-cell Translocation Gene 2 (BTG2) | | Descriptor: | 1,2-ETHANEDIOL, Protein BTG2 | | Authors: | Sampathkumar, P, Romero, R, Wasserman, S, Hu, S, Maletic, M, Freeman, J, Tarun, G, Atwell, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-23 | | Release date: | 2008-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human B-cell Translocation Gene 2 (BTG2)
To be Published
|
|
3EUM
 
 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6066 | | Descriptor: | 3,6-Bis[3-(azepan-1-yl)propionamido]acridine, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', POTASSIUM ION | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-10-10 | | Release date: | 2008-10-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
3ERU
 
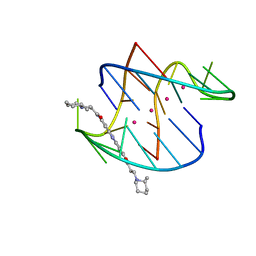 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6045 | | Descriptor: | 3,6-Bis{3-[(2R)-2-methylpiperidino)]propionamido}acridine, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', POTASSIUM ION | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-10-03 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
3EUI
 
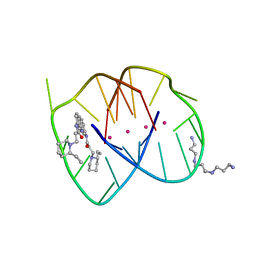 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6042 in a large unit cell | | Descriptor: | 3,6-Bis{3-[(2R)-(2-ethylpiperidino)]propionamido}acridine, 3-[(2R)-2-ethylpiperidin-1-yl]-N-[6-({3-[(2S)-2-ethylpiperidin-1-yl]propanoyl}amino)acridin-3-yl]propanamide, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', ... | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-10-10 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
3ES0
 
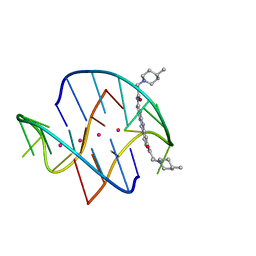 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6048 | | Descriptor: | 3,6-Bis[3-(4-methylpiperidino)propionamido]acridine, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', POTASSIUM ION | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-10-03 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
3FGC
 
 | | Crystal Structure of the Bacterial Luciferase:Flavin Complex Reveals the Basis of Intersubunit Communication | | Descriptor: | Alkanal monooxygenase alpha chain, Alkanal monooxygenase beta chain, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Campbell, Z.T, Weichsel, A, Montfort, W.R, Baldwin, T.O. | | Deposit date: | 2008-12-05 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the bacterial luciferase/flavin complex provides insight into the function of the beta subunit.
Biochemistry, 48, 2009
|
|
2NPO
 
 | | Crystal structure of putative transferase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | Acetyltransferase | | Authors: | Jin, X, Bera, A, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative transferase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
6QTI
 
 | |
6S59
 
 | | Structure of ovine transhydrogenase in the apo state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Nicotinamide nucleotide transhydrogenase | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2019-07-01 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure and mechanism of mitochondrial proton-translocating transhydrogenase.
Nature, 573, 2019
|
|
6QUE
 
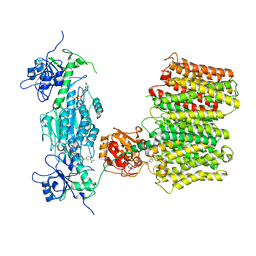 | |
6ZK9
 
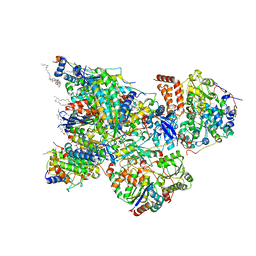 | | Peripheral domain of open complex I during turnover | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-10-07 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
2K5Z
 
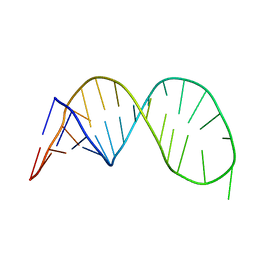 | |
5IEJ
 
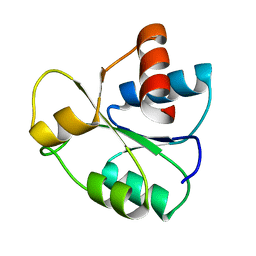 | |
5IEB
 
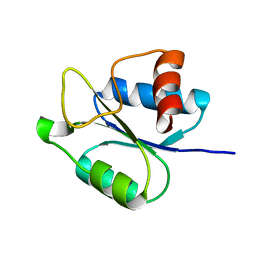 | |
6UJC
 
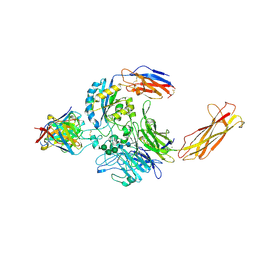 | | Integrin alpha-v beta-8 in complex with the Fabs C6-RGD3 and 11D12v2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C6-RGD3 heavy chain Fab, ... | | Authors: | Campbell, M.G, Cormier, A, Cheng, Y, Nishimura, S.L. | | Deposit date: | 2019-10-02 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Cryo-EM Reveals Integrin-Mediated TGF-beta Activation without Release from Latent TGF-beta.
Cell, 180, 2020
|
|
4WKM
 
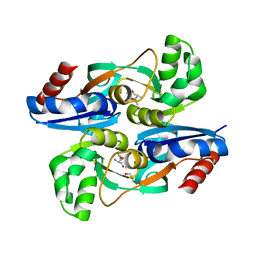 | | AmpR effector binding domain from Citrobacter freundii bound to UDP-MurNAc-pentapeptide | | Descriptor: | ALA-FGA-API-DAL-DAL, GLYCEROL, LysR family transcriptional regulator, ... | | Authors: | Vadlamani, G, Reeve, T.M, Mark, B.L. | | Deposit date: | 2014-10-02 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The beta-Lactamase Gene Regulator AmpR Is a Tetramer That Recognizes and Binds the d-Ala-d-Ala Motif of Its Repressor UDP-N-acetylmuramic Acid (MurNAc)-pentapeptide.
J.Biol.Chem., 290, 2015
|
|
6UJB
 
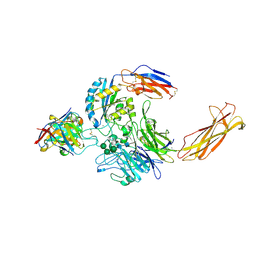 | | Integrin alpha-v beta-8 in complex with the Fabs C6D4 and 11D12v2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C6D4 heavy chain Fab, ... | | Authors: | Campbell, M.G, Cormier, A, Cheng, Y, Nishimura, S.L. | | Deposit date: | 2019-10-02 | | Release date: | 2020-02-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Cryo-EM Reveals Integrin-Mediated TGF-beta Activation without Release from Latent TGF-beta.
Cell, 180, 2020
|
|
9EKC
 
 | | cryo-EM of CL1 tube (outer) | | Descriptor: | CL1 dimer | | Authors: | Wang, F, Gnewou, O, Tuachi, A, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2024-12-02 | | Release date: | 2025-03-26 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Amphipathic Antimicrobial Peptides Illuminate a Reciprocal Relationship Between Self-assembly and Cytolytic Activity.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
6HMO
 
 | |
2Y20
 
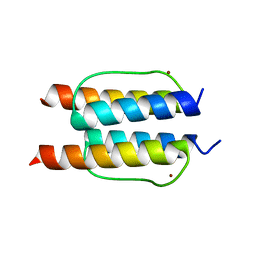 | | The mechanisms of HAMP-mediated signaling in transmembrane receptors - the A291I mutant | | Descriptor: | UNCHARACTERIZED PROTEIN, ZINC ION | | Authors: | Zeth, K, Ferris, H.U, Hulko, M, Lupas, A.N. | | Deposit date: | 2010-12-12 | | Release date: | 2011-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Mechanisms of Hamp-Mediated Signaling in Transmembrane Receptors.
Structure, 19, 2011
|
|
3LNR
 
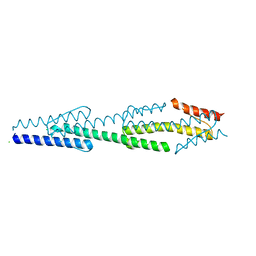 | |
5FWX
 
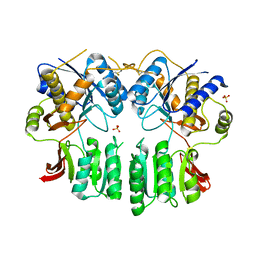 | | Crystal structure of the AMPA receptor GluA2/A4 N-terminal domain heterodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMATE RECEPTOR 2, GLUTAMATE RECEPTOR 4, ... | | Authors: | Garcia-Nafria, J, Herguedas, B, Greger, I.H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-03-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Organization of Heteromeric Ampa-Type Glutamate Receptors.
Science, 352, 2016
|
|
5FWY
 
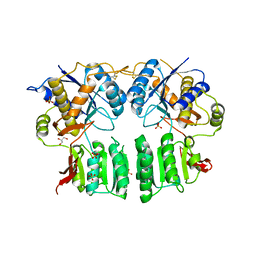 | | Crystal structure of the AMPA receptor GluA2/A3 N-terminal domain heterodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMATE RECEPTOR 2, GLUTAMATE RECEPTOR 3, ... | | Authors: | Herguedas, B, Garcia-Nafria, J, Greger, I.H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure and Organization of Heteromeric Ampa-Type Glutamate Receptors.
Science, 352, 2016
|
|
