1ULM
 
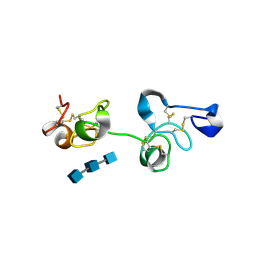 | | Crystal Structure of Pokeweed Lectin-D2 complexed with tri-N-acetylchitotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, lectin-D2 | | Authors: | Hayashida, M, Fujii, T, Ishiguro, M, Hata, Y. | | Deposit date: | 2003-09-12 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Similarity between protein-protein and protein-carbohydrate interactions, revealed by two crystal structures of lectins from the roots of pokeweed.
J.Mol.Biol., 334, 2003
|
|
2OBT
 
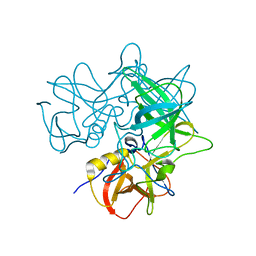 | |
3OGO
 
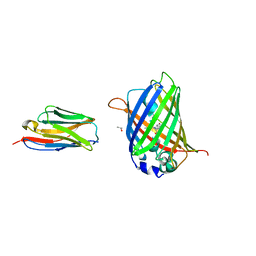 | | Structure of the GFP:GFP-nanobody complex at 2.8 A resolution in spacegroup P21212 | | Descriptor: | GFP-nanobody, Green fluorescent protein, ISOPROPYL ALCOHOL | | Authors: | Kubala, M.H, Kovtun, O, Alexandrov, K, Collins, B.M. | | Deposit date: | 2010-08-17 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and thermodynamic analysis of the GFP:GFP-nanobody complex.
Protein Sci., 19, 2010
|
|
3OED
 
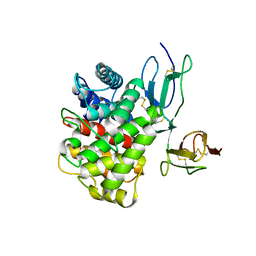 | |
3OF6
 
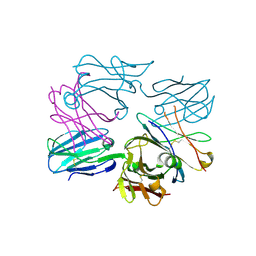 | | Human pre-T cell receptor crystal structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pre T-cell antigen receptor alpha, T cell receptor beta chain | | Authors: | Pang, S.S. | | Deposit date: | 2010-08-13 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis for autonomous dimerization of the pre-T-cell antigen receptor
Nature, 467, 2010
|
|
1P6A
 
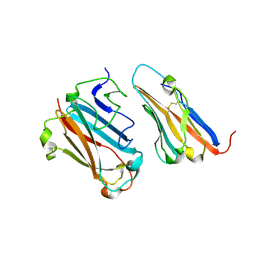 | | STRUCTURAL BASIS FOR VARIATION IN ADENOVIRUS AFFINITY FOR THE CELLULAR RECEPTOR CAR (S489Y MUTANT) | | Descriptor: | Coxsackievirus and adenovirus receptor, Fiber protein | | Authors: | Howitt, J, Bewley, M.C, Graziano, V, Flanagan, J.M, Freimuth, P. | | Deposit date: | 2003-04-29 | | Release date: | 2004-05-11 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for variation in adenovirus affinity for the cellular coxsackievirus and adenovirus receptor.
J.Biol.Chem., 278, 2003
|
|
1OT8
 
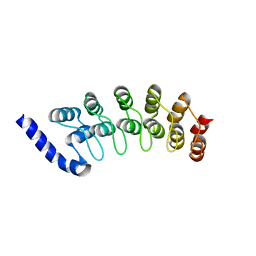 | | Structure of the Ankyrin Domain of the Drosophila Notch Receptor | | Descriptor: | MAGNESIUM ION, Neurogenic locus Notch protein | | Authors: | Zweifel, M.E, Leahy, D.J, Hughson, F.M, Barrick, D. | | Deposit date: | 2003-03-21 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and stability of the ankyrin domain of the Drosophila Notch receptor
Protein Sci., 12, 2003
|
|
2OHG
 
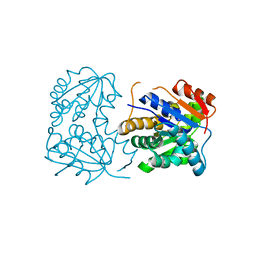 | |
1HXP
 
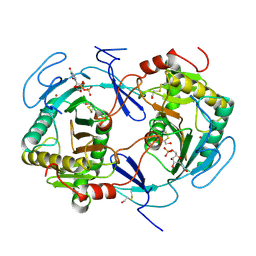 | | NUCLEOTIDE TRANSFERASE | | Descriptor: | BETA-MERCAPTOETHANOL, FE (III) ION, HEXOSE-1-PHOSPHATE URIDYLYLTRANSFERASE, ... | | Authors: | Wedekind, J.E, Frey, P.A, Rayment, I. | | Deposit date: | 1995-06-09 | | Release date: | 1996-11-08 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of galactose-1-phosphate uridylyltransferase from Escherichia coli at 1.8 A resolution.
Biochemistry, 34, 1995
|
|
1XXA
 
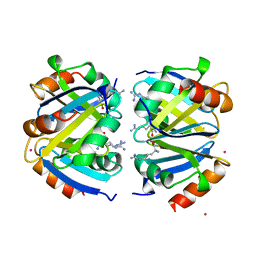 | | C-TERMINAL DOMAIN OF ESCHERICHIA COLI ARGININE REPRESSOR/ L-ARGININE COMPLEX; PB DERIVATIVE | | Descriptor: | ARGININE, ARGININE REPRESSOR, LEAD (II) ION | | Authors: | Van Duyne, G.D, Ghosh, G, Maas, W.K, Sigler, P.B. | | Deposit date: | 1995-11-03 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the oligomerization and L-arginine binding domain of the arginine repressor of Escherichia coli.
J.Mol.Biol., 256, 1996
|
|
1V3V
 
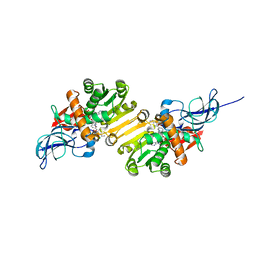 | | Crystal structure of leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase complexed with NADP and 15-oxo-PGE2 | | Descriptor: | (5E,13E)-11-HYDROXY-9,15-DIOXOPROSTA-5,13-DIEN-1-OIC ACID, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hori, T, Yokomizo, T, Ago, H, Sugahara, M, Ueno, G, Yamamoto, M, Kumasaka, T, Shimizu, T, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-06 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of leukotriene B4 12-hydroxydehydrogenase/15-Oxo-prostaglandin 13-reductase catalytic mechanism and a possible Src homology 3 domain binding loop
J.Biol.Chem., 279, 2004
|
|
1ZA7
 
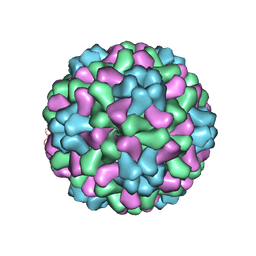 | | The crystal structure of salt stable cowpea cholorotic mottle virus at 2.7 angstroms resolution. | | Descriptor: | Coat protein | | Authors: | Bothner, B, Speir, J.A, Qu, C, Willits, D.A, Young, M.J, Johnson, J.E. | | Deposit date: | 2005-04-05 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enhanced local symmetry interactions globally stabilize a mutant virus capsid that maintains infectivity and capsid dynamics.
J.Virol., 80, 2006
|
|
1UMW
 
 | | Structure of a human Plk1 Polo-box domain/phosphopeptide complex | | Descriptor: | PEPTIDE, SERINE/THREONINE-PROTEIN KINASE PLK | | Authors: | Rellos, P, Elia, A, Yaffe, M.B, Smerdon, S.J. | | Deposit date: | 2003-08-29 | | Release date: | 2003-10-16 | | Last modified: | 2017-02-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Molecular Basis for Phosphodependent Substrate Targeting and Regulation of Plks by the Polo-Box Domain
Cell(Cambridge,Mass.), 115, 2003
|
|
1UL1
 
 | | Crystal structure of the human FEN1-PCNA complex | | Descriptor: | Flap endonuclease-1, MAGNESIUM ION, Proliferating cell nuclear antigen | | Authors: | Sakurai, S, Kitano, K, Yamaguchi, H, Hamada, K, Okada, K, Fukuda, K, Uchida, M, Ohtsuka, E, Morioka, H, Hakoshima, T. | | Deposit date: | 2003-09-05 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for recruitment of human flap endonuclease 1 to PCNA
EMBO J., 24, 2005
|
|
1ULK
 
 | | Crystal Structure of Pokeweed Lectin-C | | Descriptor: | lectin-C | | Authors: | Hayashida, M, Fujii, T, Ishiguro, M, Hata, Y. | | Deposit date: | 2003-09-12 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Similarity between protein-protein and protein-carbohydrate interactions, revealed by two crystal structures of lectins from the roots of pokeweed.
J.Mol.Biol., 334, 2003
|
|
1JLK
 
 | | Crystal structure of the Mn(2+)-bound form of response regulator Rcp1 | | Descriptor: | MANGANESE (II) ION, Response regulator RCP1, SULFATE ION | | Authors: | Im, Y.J, Rho, S.-H, Park, C.-M, Yang, S.-S, Kang, J.-G, Lee, J.Y, Song, P.-S, Eom, S.H. | | Deposit date: | 2001-07-16 | | Release date: | 2002-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a cyanobacterial phytochrome response regulator.
Protein Sci., 11, 2002
|
|
3Q4F
 
 | | Crystal structure of xrcc4/xlf-cernunnos complex | | Descriptor: | DNA repair protein XRCC4, Non-homologous end-joining factor 1 | | Authors: | Ropars, V, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2010-12-23 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structural characterization of filaments formed by human Xrcc4-Cernunnos/XLF complex involved in nonhomologous DNA end-joining.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4KS9
 
 | | Crystal Structure of Malonyl-CoA decarboxylase (Rmet_2797) from Cupriavidus metallidurans, Northeast Structural Genomics Consortium Target CrR76 | | Descriptor: | MAGNESIUM ION, Malonyl-CoA decarboxylase | | Authors: | Forouhar, F, Tran, T.H, Lew, S, Seetharaman, J, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-17 | | Release date: | 2013-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of malonyl-coenzyme a decarboxylase provide insights into its catalytic mechanism and disease-causing mutations.
Structure, 21, 2013
|
|
2WBO
 
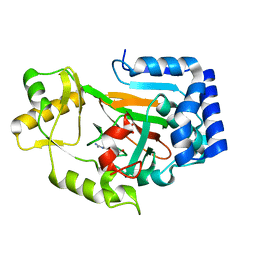 | | Crystal structure of VioC in complex with L-arginine | | Descriptor: | ARGININE, FE (II) ION, L(+)-TARTARIC ACID, ... | | Authors: | Helmetag, V, Samel, S.A, Thomas, M.G, Marahiel, M.A, Essen, L.-O. | | Deposit date: | 2009-03-02 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Basis for the Erythro-Stereospecificity of the L-Arginine Oxygenase Vioc in Viomycin Biosynthesis.
FEBS J., 276, 2009
|
|
3PWA
 
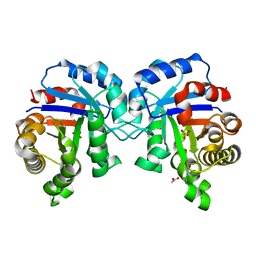 | | Structure of C126A mutant of Plasmodium falciparum triosephosphate isomerase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Samanta, M, Banerjee, M, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2010-12-08 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Probing the role of the fully conserved Cys126 in triosephosphate isomerase by site-specific mutagenesis--distal effects on dimer stability.
Febs J., 278, 2011
|
|
3PVF
 
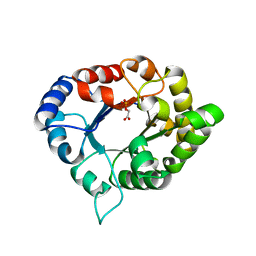 | | Structure of C126S mutant of Plasmodium falciparum triosephosphate isomerase complexed with PGA | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Samanta, M, Banerjee, M, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2010-12-07 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Probing the role of the fully conserved Cys126 in triosephosphate isomerase by site-specific mutagenesis--distal effects on dimer stability.
Febs J., 278, 2011
|
|
1V3C
 
 | | Structure of the hemagglutinin-neuraminidase from human parainfluenza virus type III: complex with NEU5AC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lawrence, M.C, Borg, N.A, Streltsov, V.A, Pilling, P.A, Epa, V.C, Varghese, J.N, McKimm-Breschkin, J.L, Colman, P.M. | | Deposit date: | 2003-10-30 | | Release date: | 2004-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Haemagglutinin-neuraminidase from Human Parainfluenza Virus Type III
J.Mol.Biol., 335, 2004
|
|
1MNU
 
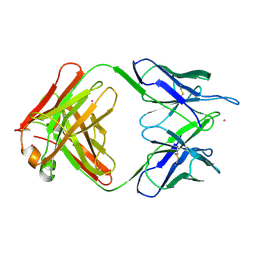 | | UNLIGANDED BACTERICIDAL ANTIBODY AGAINST NEISSERIA MENINGITIDIS | | Descriptor: | CADMIUM ION, PROTEIN (IGG2A-KAPPA ANTIBODY MN12H2 (HEAVY CHAIN)), PROTEIN (IGG2A-KAPPA ANTIBODY MN12H2 (LIGHT CHAIN)) | | Authors: | Van Den Elsen, J, Vandeputte-Rutten, L, Kroon, J, Gros, P. | | Deposit date: | 1999-04-30 | | Release date: | 1999-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bactericidal antibody recognition of meningococcal PorA by induced fit. Comparison of liganded and unliganded Fab structures.
J.Biol.Chem., 274, 1999
|
|
1TQE
 
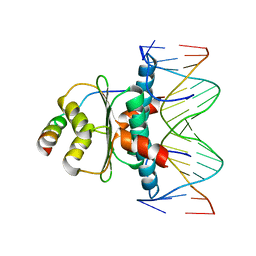 | | Mechanism of recruitment of class II histone deacetylases by myocyte enhancer factor-2 | | Descriptor: | Histone deacetylase 9, MEF2 binding site of nur77 promoter, Myocyte-specific enhancer factor 2B | | Authors: | Chen, L, Han, A, He, J, Wu, Y, Liu, J.O. | | Deposit date: | 2004-06-17 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of Recruitment of Class II Histone Deacetylases by Myocyte Enhancer Factor-2.
J.Mol.Biol., 345, 2005
|
|
7F3P
 
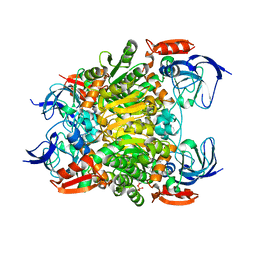 | | Crystal structure of a nadp-dependent alcohol dehydrogenase mutant in apo form | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent isopropanol dehydrogenase, ZINC ION | | Authors: | Han, X, Bi, Y, Wei, H.L, Gao, J, Li, Q, Qu, G, Sun, Z.T, Liu, W.D. | | Deposit date: | 2021-06-16 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Unlocking the Stereoselectivity and Substrate Acceptance of Enzymes: Proline-Induced Loop Engineering Test.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
