5NIQ
 
 | |
5NCA
 
 | | Solution structure of ComGC from Streptococcus pneumoniae | | Descriptor: | Competence protein ComGC | | Authors: | Erlendsson, S, Schmeider, P, Lichtenberg, C, Teilum, K, Akbey, U. | | Deposit date: | 2017-03-03 | | Release date: | 2017-07-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of the competence pilus major pilin ComGC in Streptococcus pneumoniae.
J. Biol. Chem., 292, 2017
|
|
5YOF
 
 | |
5ODM
 
 | | NtiPr polyamide in complex with 5'CGATGTACTACG3 | | Descriptor: | 3-(3-azaniumylpropanoylamino)propyl-dimethyl-azanium, 4-AMINO-(1-METHYLIMIDAZOLE)-2-CARBOXYLIC ACID, 4-AMINO-(1-METHYLPYRROLE)-2-CARBOXYLIC ACID, ... | | Authors: | Padroni, G, Parkinson, J, Burley, G.A. | | Deposit date: | 2017-07-06 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA duplex distortion induced by thiazole-containing hairpin polyamides.
Nucleic Acids Res., 46, 2018
|
|
5OAK
 
 | |
5YOD
 
 | |
5NQH
 
 | | Structure of the human Fe65-PTB2 homodimer | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 1, GLYCEROL, SULFATE ION | | Authors: | Feilen, L.P, Haubrich, K, Sinning, I, Konietzko, U, Kins, S, Simon, B, Wild, K. | | Deposit date: | 2017-04-20 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fe65-PTB2 Dimerization Mimics Fe65-APP Interaction.
Front Mol Neurosci, 10, 2017
|
|
7JX7
 
 | | BRD2-BD2 in complex with a diacetylated-H2A.Z peptide | | Descriptor: | Bromodomain-containing protein 2, Diacetylated-H2A.Z peptide | | Authors: | Patel, K, Low, J.K.K, Mackay, J.P, Solomon, P.D. | | Deposit date: | 2020-08-26 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The bromodomains of BET family proteins can recognize diacetylated histone H2A.Z.
Protein Sci., 30, 2021
|
|
5WUZ
 
 | | Morintides mO1 | | Descriptor: | Morintide mO1 | | Authors: | Xiao, T, Tam, J.P. | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Morintides: 8C-Hevein-like peptides without a protein cargo from the drumstick tree Moringa oleifera
To Be Published
|
|
5ZEW
 
 | |
5WYO
 
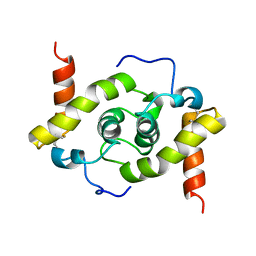 | | Solution structure of E.coli HdeA | | Descriptor: | Acid stress chaperone HdeA | | Authors: | Yang, C, Hu, Y, Jin, C. | | Deposit date: | 2017-01-14 | | Release date: | 2017-11-22 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Characterizations of the Interactions between Escherichia coli Periplasmic Chaperone HdeA and Its Native Substrates during Acid Stress
Biochemistry, 56, 2017
|
|
5WXE
 
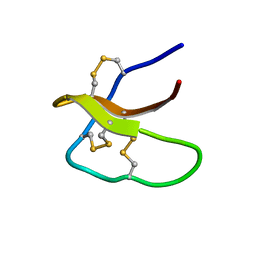 | | Highly disulfide-constrained antifeedant jasmintides from Jasminum sambac flowers | | Descriptor: | jasmintide js3 | | Authors: | Kumari, G, Wong, K.H, Serra, A, Shin, J, Yoon, H.S, Sze, S.K, James, P.T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-01-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Highly disulfide-constrained antifeedant jasmintides from Jasminum sambac flowers
To Be Published
|
|
5WPY
 
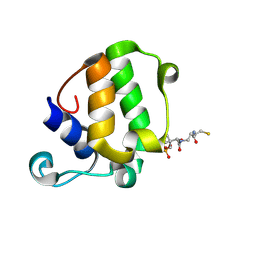 | |
5OWJ
 
 | | The dynamic dimer structure of the chaperone Trigger Factor (conformer 2) | | Descriptor: | Trigger factor | | Authors: | Morgado, L, Burmann, B.M, Sharpe, T, Mazur, A, Hiller, S. | | Deposit date: | 2017-09-01 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The dynamic dimer structure of the chaperone Trigger Factor.
Nat Commun, 8, 2017
|
|
5NWX
 
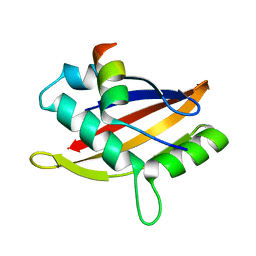 | | Insight into the molecular recognition mechanism of the coactivator NCoA1 by STAT6 | | Descriptor: | Nuclear receptor coactivator 1, Signal transducer and activator of transcription 6 | | Authors: | Russo, L, Giller, K, Pfitzner, E, Griesinger, C, Becker, S. | | Deposit date: | 2017-05-08 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Insight into the molecular recognition mechanism of the coactivator NCoA1 by STAT6.
Sci Rep, 7, 2017
|
|
5OCZ
 
 | | Free DNA_hairpin polyamides studies | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*GP*TP*AP*CP*AP*TP*CP*G)-3') | | Authors: | Padroni, G, Parkinson, J, Burley, G.A. | | Deposit date: | 2017-07-04 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA duplex distortion induced by thiazole-containing hairpin polyamides.
Nucleic Acids Res., 46, 2018
|
|
2RNQ
 
 | |
2M36
 
 | |
8WRG
 
 | |
9CRB
 
 | | Structure of Avt1, a novel peptide from the sea anemone Aulactinia veratra | | Descriptor: | Avt1 peptide | | Authors: | Albar, R, Sanches, K, Wai, D, Norton, R. | | Deposit date: | 2024-07-22 | | Release date: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure and functional studies of Avt1, a novel peptide from the sea anemone Aulactinia veratra.
Biochim Biophys Acta Proteins Proteom, 1873, 2024
|
|
1PIS
 
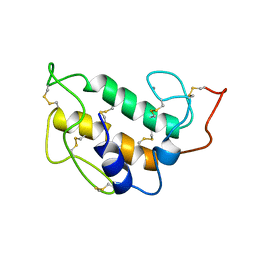 | | SOLUTION STRUCTURE OF PORCINE PANCREATIC PHOSPHOLIPASE A2 | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Van Den Berg, B.D, Tessari, M, De Haas, G.H, Verheij, H.M, Boelens, R, Kaptein, R. | | Deposit date: | 1994-12-22 | | Release date: | 1995-06-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine pancreatic phospholipase A2.
EMBO J., 14, 1995
|
|
1PIR
 
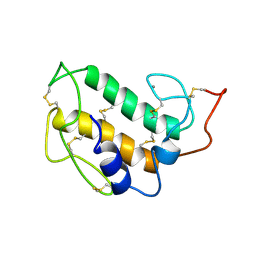 | | SOLUTION STRUCTURE OF PORCINE PANCREATIC PHOSPHOLIPASE A2 | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Van Den Berg, B.D, Tessari, M, De Haas, G.H, Verheij, H.M, Boelens, R, Kaptein, R. | | Deposit date: | 1994-12-22 | | Release date: | 1995-06-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine pancreatic phospholipase A2.
EMBO J., 14, 1995
|
|
2NOC
 
 | | Solution Structure of Putative periplasmic protein: Northest Structural Genomics Target StR106 | | Descriptor: | Putative periplasmic protein | | Authors: | Zhang, Q, Liu, G, Wang, H, Nwosu, C, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Putative periplasmic protein: Northest Structural Genomics Target StR106
To be Published
|
|
6U4N
 
 | |
1QO6
 
 | |
