1FVQ
 
 | | SOLUTION STRUCTURE OF THE YEAST COPPER TRANSPORTER DOMAIN CCC2A IN THE APO AND CU(I) LOADED STATES | | Descriptor: | COPPER-TRANSPORTING ATPASE | | Authors: | Banci, L, Bertini, I, Ciofi Baffoni, S, Huffman, D.L, O'Halloran, T.V. | | Deposit date: | 2000-09-20 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the yeast copper transporter domain Ccc2a in the apo and Cu(I)-loaded states.
J.Biol.Chem., 276, 2001
|
|
1IB9
 
 | |
1IXT
 
 | | Structure of a Novel P-Superfamily Spasmodic Conotoxin Reveals an Inhibitory Cystine Knot Motif | | Descriptor: | spasmodic protein tx9a-like protein | | Authors: | Miles, L.A, Dy, C.Y, Nielsen, J, Barnham, K.J, Hinds, M.G, Olivera, B.M, Bulaj, G, Norton, R.S. | | Deposit date: | 2002-07-04 | | Release date: | 2003-01-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of a Novel P-Superfamily Spasmodic Conotoxin Reveals an Inhibitory Cystine Knot Motif
J.Biol.Chem., 277, 2002
|
|
1EI2
 
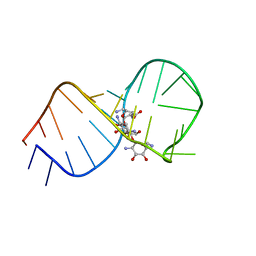 | | STRUCTURAL BASIS FOR RECOGNITION OF THE RNA MAJOR GROOVE IN THE TAU EXON 10 SPLICING REGULATORY ELEMENT BY AMINOGLYCOSIDE ANTIBIOTICS | | Descriptor: | NEOMYCIN, TAU EXON 10 SRE RNA | | Authors: | Varani, L, Spillantini, M.G, Goedert, M, Varani, G. | | Deposit date: | 2000-02-23 | | Release date: | 2000-03-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of the RNA major groove in the tau exon 10 splicing regulatory element by aminoglycoside antibiotics.
Nucleic Acids Res., 28, 2000
|
|
1HP3
 
 | |
1HYJ
 
 | |
1HYI
 
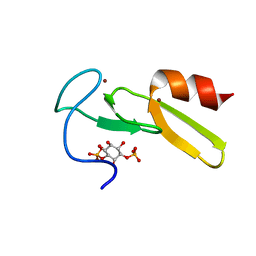 | | SOLUTION STRUCTURE OF THE EEA1 FYVE DOMAIN COMPLEXED WITH INOSITOL 1,3-BISPHOSPHATE | | Descriptor: | ENDOSOME-ASSOCIATED PROTEIN, PHOSPHORIC ACID MONO-(2,3,4,6-TETRAHYDROXY-5-PHOSPHONOOXY-CYCLOHEXYL) ESTER, ZINC ION | | Authors: | Kutateladze, T, Overduin, M. | | Deposit date: | 2001-01-19 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural mechanism of endosome docking by the FYVE domain.
Science, 291, 2001
|
|
1HZ5
 
 | | CRYSTAL STRUCTURES OF THE B1 DOMAIN OF PROTEIN L FROM PEPTOSTREPTOCOCCUS MAGNUS, WITH A TYROSINE TO TRYPTOPHAN SUBSTITUTION | | Descriptor: | PROTEIN L, ZINC ION | | Authors: | O'Neill, J.W, Kim, D.E, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-01-23 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the B1 domain of protein L from Peptostreptococcus magnus with a tyrosine to tryptophan substitution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HZ6
 
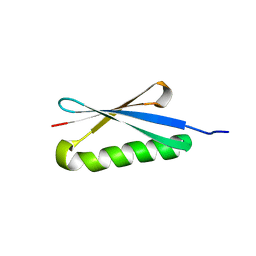 | | CRYSTAL STRUCTURES OF THE B1 DOMAIN OF PROTEIN L FROM PEPTOSTREPTOCOCCUS MAGNUS WITH A TYROSINE TO TRYPTOPHAN SUBSTITUTION | | Descriptor: | PROTEIN L | | Authors: | O'Neill, J.W, Kim, D.E, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-01-23 | | Release date: | 2001-04-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the B1 domain of protein L from Peptostreptococcus magnus with a tyrosine to tryptophan substitution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1IEN
 
 | | SOLUTION STRUCTURE OF TIA | | Descriptor: | PROTEIN TIA | | Authors: | Sharpe, I.A, Gehrmann, J, Loughnan, M.L, Thomas, L, Adams, D.A, Atkins, A, Palant, E, Craik, D.J, Adams, D.J, Alewood, P.F, Lewis, R.J. | | Deposit date: | 2001-04-10 | | Release date: | 2002-04-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Two new classes of conopeptides inhibit the alpha1-adrenoceptor and noradrenaline transporter.
Nat.Neurosci., 4, 2001
|
|
1IFW
 
 | | SOLUTION STRUCTURE OF C-TERMINAL DOMAIN OF POLY(A) BINDING PROTEIN FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | POLYADENYLATE-BINDING PROTEIN, CYTOPLASMIC AND NUCLEAR | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Sprules, T, Ekiel, I, Gehring, K. | | Deposit date: | 2001-04-13 | | Release date: | 2002-07-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the orphan PABC domain from Saccharomyces cerevisiae poly(A)-binding protein.
J.Biol.Chem., 277, 2002
|
|
1IV0
 
 | |
1IGV
 
 | | BOVINE CALBINDIN D9K BINDING MN2+ | | Descriptor: | MANGANESE (II) ION, VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN, INTESTINAL | | Authors: | Andersson, E.M. | | Deposit date: | 2001-04-18 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the negative allostery between Ca(2+)- and Mg(2+)-binding in the intracellular Ca(2+)-receptor calbindin D9k.
Protein Sci., 6, 1997
|
|
1IU2
 
 | | The first PDZ domain of PSD-95 | | Descriptor: | PSD-95 | | Authors: | Long, J.-F, Tochio, H, Wang, P, Sala, C, Niethammer, M, Sheng, M, Zhang, M. | | Deposit date: | 2002-02-19 | | Release date: | 2003-03-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Supramodular structure and synergistic target binding of the N-terminal tandem PDZ domains of PSD-95
J.MOL.BIOL., 327, 2003
|
|
1IUY
 
 | |
6GIF
 
 | |
6K50
 
 | |
5IPO
 
 | |
7KLO
 
 | |
7KLR
 
 | |
2W84
 
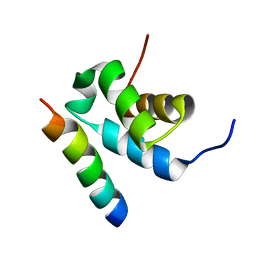 | | Structure of Pex14 in complex with Pex5 | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PEX14, PEROXISOMAL TARGETING SIGNAL 1 RECEPTOR | | Authors: | Neufeld, C, Filipp, F.V, Simon, B, Neuhaus, A, Schueller, N, David, C, Kooshapur, H, Madl, T, Erdmann, R, Schliebs, W, Wilmanns, M, Sattler, M. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for competitive interactions of Pex14 with the import receptors Pex5 and Pex19.
EMBO J., 28, 2009
|
|
2W85
 
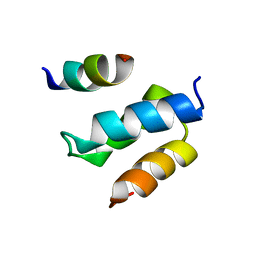 | | Structure of Pex14 in complex with Pex19 | | Descriptor: | PEROXIN-19, PEROXISOMAL MEMBRANE ANCHOR PROTEIN PEX14 | | Authors: | Neufeld, C, Filipp, F.V, Simon, B, Neuhaus, A, Schueller, N, David, C, Kooshapur, H, Madl, T, Erdmann, R, Schliebs, W, Wilmanns, M, Sattler, M. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Competitive Interactions of Pex14 with the Import Receptors Pex5 and Pex19.
Embo J., 28, 2009
|
|
6KHV
 
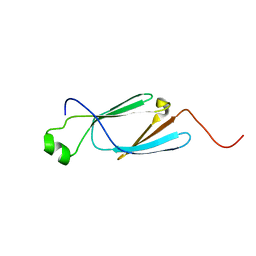 | | Solution Structure of the CS2 Domain of USP19 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 19 | | Authors: | Xue, W, Hu, H.Y. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Domain interactions reveal auto-inhibition of the deubiquitinating enzyme USP19 and its activation by HSP90 in the modulation of huntingtin aggregation.
Biochem.J., 477, 2020
|
|
6KG8
 
 | | Solution structure of CaCohA2 from Clostridium acetobutylicum | | Descriptor: | Probably cellulosomal scaffolding protein, secreted cellulose-binding and cohesin domain | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
6K3K
 
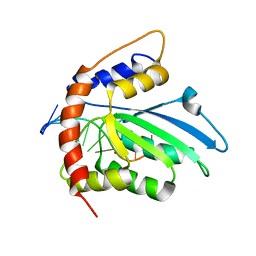 | | Solution structure of APOBEC3G-CD2 with ssDNA, Product B | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, DNA/RNA (5'-D(*AP*TP*TP*CP*UP*(ICY)P*AP*AP*TP*T)-3'), ZINC ION | | Authors: | Cao, C, Yan, X, Lan, W, Wang, C. | | Deposit date: | 2019-05-19 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Investigations on the Interactions between Cytidine Deaminase Human APOBEC3G and DNA.
Chem Asian J, 14, 2019
|
|
