4UI5
 
 | | Crystal structure of human tankyrase 2 in complex with TA-41 | | Descriptor: | 8-methoxy-2-(4-methylphenyl)-3,4-dihydroquinazolin-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-03-27 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Activity Relationships of 2-Arylquinazolin-4-Ones as Highly Selective and Potent Inhibitors of the Tankyrases.
Eur.J.Med.Chem., 118, 2016
|
|
4HK6
 
 | |
4V7F
 
 | | Arx1 pre-60S particle. | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Leidig, C, Thoms, M, Holdermann, I, Bradatsch, B, Berninghausen, O, Bange, G, Sinning, I, Hurt, E, Beckmann, R. | | Deposit date: | 2013-12-10 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle.
Nat Commun, 5, 2014
|
|
4V2B
 
 | | rat Unc5D Ig domain 1 | | Descriptor: | PROTEIN UNC5D | | Authors: | Seiradake, E, del Toro, D, Nagel, D, Cop, F, Haertl, R, Ruff, T, Seyit-Bremer, G, Harlos, K, Border, E.C, Acker-Palmer, A, Jones, E.Y, Klein, R. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-05 | | Last modified: | 2015-07-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flrt Structure: Balancing Repulsion and Cell Adhesion in Cortical and Vascular Development
Neuron, 84, 2014
|
|
8JFK
 
 | | PhK holoenzyme in inactive state, muscle isoform | | Descriptor: | Calmodulin-1, FARNESYL, Phosphorylase b kinase gamma catalytic chain, ... | | Authors: | Yang, X.K, Xiao, J.Y. | | Deposit date: | 2023-05-18 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Architecture and activation of human muscle phosphorylase kinase.
Nat Commun, 15, 2024
|
|
4V0M
 
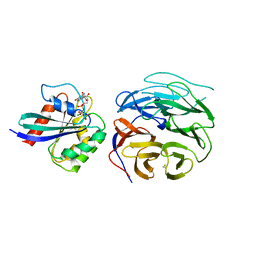 | | Crystal structure of BBS1N in complex with ARL6DN | | Descriptor: | ARF-LIKE SMALL GTPASE, BARDET-BIEDL SYNDROME 1 PROTEIN, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mourao, A, Lorentzen, E. | | Deposit date: | 2014-09-17 | | Release date: | 2014-11-19 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural Basis for Membrane Targeting of the Bbsome by Arl6
Nat.Struct.Mol.Biol., 21, 2014
|
|
4V6C
 
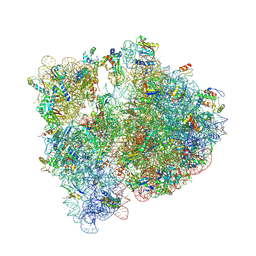 | | Crystal structure of the E. coli 70S ribosome in an intermediate state of ratcheting | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, W, Dunkle, J.A, Cate, J.H.D. | | Deposit date: | 2009-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structures of the ribosome in intermediate States of ratcheting.
Science, 325, 2009
|
|
4E7P
 
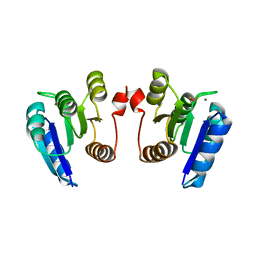 | | Crystal structure of receiver domain of putative NarL family response regulator spr1814 from Streptococcus pneumoniae in the presence of the phosphoryl analog beryllofluoride | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Response regulator | | Authors: | Park, A.K, Moon, J.H, Lee, K.S, Chi, Y.M. | | Deposit date: | 2012-03-18 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Crystal structure of receiver domain of putative NarL family response regulator spr1814 from Streptococcus pneumoniae in the absence and presence of the phosphoryl analog beryllofluoride.
Biochem.Biophys.Res.Commun., 421, 2012
|
|
4HMZ
 
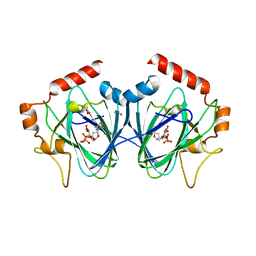 | | Crystal Structure of ChmJ, a 3'-monoepimerase from Streptomyces bikiniensis in complex with dTDP-quinovose | | Descriptor: | 1,2-ETHANEDIOL, Putative 3-epimerase in D-allose pathway, [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl (2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-methyltetrahydro-2H-pyran-2-yl dihydrogen diphosphate | | Authors: | Holden, H.M, Kubiak, R.L. | | Deposit date: | 2012-10-18 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Studies on a 3'-Epimerase Involved in the Biosynthesis of dTDP-6-deoxy-d-allose.
Biochemistry, 51, 2012
|
|
4W2H
 
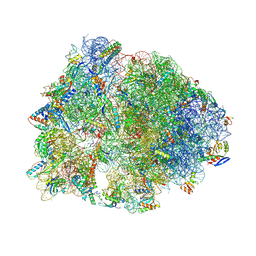 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with pactamycin (co-crystallized), mRNA and deacylated tRNA in the P site | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Osterman, I.A, Szal, T, Tashlitsky, V.N, Serebryakova, M.V, Kusochek, P, Bulkley, D, Malanicheva, I.A, Efimenko, T.A, Efremenkova, O.V, Konevega, A.L, Shaw, K.J, Bogdanov, A.A, Rodnina, M.V, Dontsova, O.A, Mankin, A.S, Steitz, T.A, Sergiev, P.V. | | Deposit date: | 2014-09-12 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Amicoumacin a inhibits translation by stabilizing mRNA interaction with the ribosome.
Mol.Cell, 56, 2014
|
|
4UR6
 
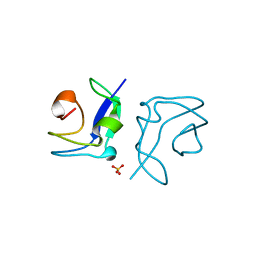 | | Structure of the type III fish antifreeze protein from Zoarces viviparus ZvAFP6 | | Descriptor: | SULFATE ION, TYPE III ANTIFREEZE PROTEIN 6 | | Authors: | Wilkens, C, Poulsen, J.-C.N, Ramloev, H, Lo Leggio, L. | | Deposit date: | 2014-06-26 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Purification, Crystal Structure Determination and Functional Characterization of Type III Antifreeze Proteins from the European Eelpout Zoarces Viviparus.
Cryobiology, 69, 2014
|
|
4HK7
 
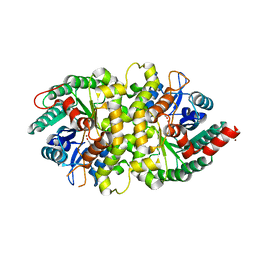 | | Crystal structure of Cordyceps militaris IDCase in complex with uracil | | Descriptor: | URACIL, Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Zhu, J, Ding, J. | | Deposit date: | 2012-10-15 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4UUR
 
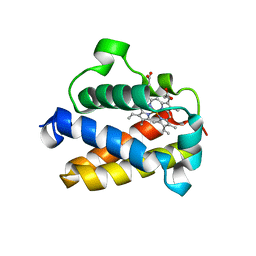 | | Cold-adapted truncated hemoglobin from the Antarctic marine bacterium Pseudoalteromonas haloplanktis TAC125 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE HEMOGLOBIN-LIKE OXYGEN-BINDING PROTEIN | | Authors: | Pesce, A, Giordano, D, Riccio, A, Nardini, M, Caldelli, E, Howes, B, Bustamante, J.P, Boechi, L, Estrin, D, di Prisco, G, Smulevich, G, Verde, C, Bolognesi, M. | | Deposit date: | 2014-07-31 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Flexibility of the Heme Cavity in the Cold-Adapted Truncated Hemoglobin from the Antarctic Marine Bacterium Pseudoalteromonas Haloplanktis Tac125.
FEBS J., 282, 2015
|
|
4HK5
 
 | | Crystal structure of Cordyceps militaris IDCase in apo form | | Descriptor: | Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Zhu, J, Ding, J. | | Deposit date: | 2012-10-15 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4V0T
 
 | | Monomeric pseudorabies virus protease pUL26N at 2.1 A resolution | | Descriptor: | UL26 | | Authors: | Zuehlsdorf, M, Werten, S, Palm, G.J, Hinrichs, W. | | Deposit date: | 2014-09-18 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin Pul26N, a Herpesvirus Serine Protease
Plos Pathog., 11, 2015
|
|
4V4A
 
 | | Crystal Structure of the Wild Type Ribosome from E. Coli 70S Ribosome. | | Descriptor: | 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, 30S ribosomal protein S10, ... | | Authors: | Vila-Sanjurjo, A, Ridgeway, W.K, Seymaner, V, Zhang, W, Santoso, S, Yu, K, Cate, J.H.D. | | Deposit date: | 2003-06-13 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (9.5 Å) | | Cite: | X-ray crystal structures of the WT and a hyper-accurate ribosome from Escherichia coli
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4V4T
 
 | | Crystal structure of the whole ribosomal complex with a stop codon in the A-site. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2005-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.46 Å) | | Cite: | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
4V55
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with gentamicin and ribosome recycling factor (RRF). | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 16S rRNA, 23S rRNA, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-17 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4WAK
 
 | |
4HN0
 
 | |
5MGD
 
 | | STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 6-Galactosyl-lactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Rico-Diaz, A, Ramirez-Escudero, M, Vizoso Vazquez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2016-11-21 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural features of Aspergillus niger beta-galactosidase define its activity against glycoside linkages.
FEBS J., 284, 2017
|
|
2YK5
 
 | | Structure of Neisseria LOS-specific sialyltransferase (NST), in complex with CMP. | | Descriptor: | 1,2-ETHANEDIOL, CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE-ALPHA-2,3-SIALYLTRANSFERASE, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Lin, L.Y.C, Rakic, B, Chiu, C.P.C, Lameignere, E, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2011-05-25 | | Release date: | 2011-08-31 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and Mechanism of the Lipooligosaccharide Sialyltransferase from Neisseria Meningitidis
J.Biol.Chem., 286, 2011
|
|
4WAM
 
 | |
4WB9
 
 | | Human ALDH1A1 complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, Retinal dehydrogenase 1, ... | | Authors: | Morgan, C.A, Hurley, T.D. | | Deposit date: | 2014-09-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Development of a high-throughput in vitro assay to identify selective inhibitors for human ALDH1A1.
Chem.Biol.Interact., 234, 2015
|
|
4WCW
 
 | | Ribosomal silencing factor during starvation or stationary phase (RsfS) from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, Ribosomal silencing factor RsfS | | Authors: | Li, X, Sun, Q, Jiang, C, Yang, K, Hung, L, Zhang, J, Sacchettini, J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-09-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Ribosomal Silencing Factor Bound to Mycobacterium tuberculosis Ribosome.
Structure, 23, 2015
|
|
