3TW3
 
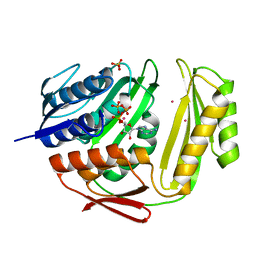 | | Crystal structure of RtcA.ATP.Co ternary complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, COBALT (II) ION, RNA 3'-terminal phosphate cyclase, ... | | Authors: | Chakravarty, A.K, Smith, P, Shuman, S. | | Deposit date: | 2011-09-21 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of RNA 3'-phosphate cyclase bound to ATP reveal the mechanism of nucleotidyl transfer and metal-assisted catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5EYV
 
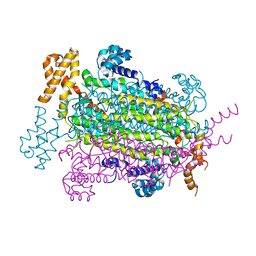 | | Crystal Structure of Adenylosuccinate lyase from Schistosoma mansoni in APO form. | | Descriptor: | Adenylosuccinate lyase | | Authors: | Romanello, L, Torini, J.R, Bird, L.E, Nettleship, J.E, Owens, R.J, Reddivari, Y, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2015-11-25 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural and kinetic analysis of Schistosoma mansoni Adenylosuccinate Lyase (SmADSL).
Mol. Biochem. Parasitol., 214, 2017
|
|
3L2P
 
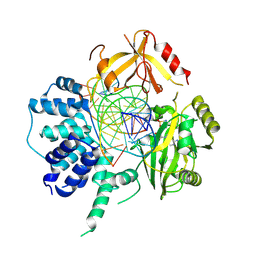 | | Human DNA Ligase III Recognizes DNA Ends by Dynamic Switching Between Two DNA Bound States | | Descriptor: | 5'-D(*GP*CP*CP*AP*GP*TP*CP*CP*GP*AP*CP*GP*AP*CP*GP*CP*AP*TP*CP*CP*CP*G)-3', 5'-D(*GP*TP*CP*GP*GP*AP*CP*TP*G)-3', 5'-D(P*CP*GP*GP*GP*AP*TP*GP*CP*GP*TP*C)-3', ... | | Authors: | Cotner-Gohara, E.A, Kim, I.K, Hammel, M, Tainer, J.A, Tomkinson, A, Ellenberger, T. | | Deposit date: | 2009-12-15 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human DNA Ligase III Recognizes DNA Ends by Dynamic Switching between Two DNA-Bound States.
Biochemistry, 49, 2010
|
|
4GXR
 
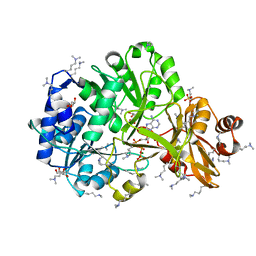 | | Structure of ATP bound RpMatB-BxBclM chimera B3 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, GLYCEROL, ... | | Authors: | Rank, K.C, Crosby, H.A, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-09-04 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the Substrate Specificity of the Rhodopseudomonas palustris Protein Acetyltransferase RpPat: IDENTIFICATION OF A LOOP CRITICAL FOR RECOGNITION BY RpPat.
J.Biol.Chem., 287, 2012
|
|
4GXQ
 
 | | Crystal Structure of ATP bound RpMatB-BxBclM chimera B1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, MAGNESIUM ION, ... | | Authors: | Rank, K.C, Crosby, H.A, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-09-04 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the Substrate Specificity of the Rhodopseudomonas palustris Protein Acetyltransferase RpPat: IDENTIFICATION OF A LOOP CRITICAL FOR RECOGNITION BY RpPat.
J.Biol.Chem., 287, 2012
|
|
1DGS
 
 | | CRYSTAL STRUCTURE OF NAD+-DEPENDENT DNA LIGASE FROM T. FILIFORMIS | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA LIGASE, ZINC ION | | Authors: | Lee, J.Y, Chang, C, Song, H.K, Kwon, S.T, Suh, S.W. | | Deposit date: | 1999-11-25 | | Release date: | 2000-11-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications.
EMBO J., 19, 2000
|
|
4R1L
 
 | |
4X45
 
 | |
4X44
 
 | | Crystal Structure of Mutant R89Q of human Adenine phosphoribosyltransferase | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenine phosphoribosyltransferase, GLYCEROL, ... | | Authors: | Pimenta, A, Pereira, H.M, Mercaldi, G, Thiemann, O.H. | | Deposit date: | 2014-12-02 | | Release date: | 2015-12-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.0535 Å) | | Cite: | Crystal Structure of Mutant R89Q of human Adenine phosphoribosyltransferase
To Be Published
|
|
3N8H
 
 | | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis | | Descriptor: | ACETIC ACID, ADENOSINE MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Maltseva, N, Kim, Y, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-28 | | Release date: | 2010-06-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis
To be Published, 2010
|
|
4RVN
 
 | |
8B4R
 
 | | Antimicrobial peptide capitellacin from polychaeta Capitella teleta in DPC (dodecylphosphocholine) micelles, monomeric form | | Descriptor: | BRICHOS domain-containing protein | | Authors: | Mironov, P.A, Reznikova, O.V, Paramonov, A.S, Shenkarev, Z.O. | | Deposit date: | 2022-09-21 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Dimerization of the beta-Hairpin Membrane-Active Cationic Antimicrobial Peptide Capitellacin from Marine Polychaeta: An NMR Structural and Thermodynamic Study.
Biomolecules, 14, 2024
|
|
1EFR
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH THE PEPTIDE ANTIBIOTIC EFRAPEPTIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE SUBUNIT ALPHA, BOVINE MITOCHONDRIAL F1-ATPASE SUBUNIT BETA, ... | | Authors: | Abrahams, J.P, Buchanan, S.K, Van Raaij, M.J, Fearnley, I.M, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 1996-05-24 | | Release date: | 1997-02-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Structure of Bovine F1-ATPase Complexed with the Peptide Antibiotic Efrapeptin.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
3NI2
 
 | | Crystal structures and enzymatic mechanisms of a Populus tomentosa 4-coumarate:CoA ligase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-coumarate:CoA ligase, 5'-O-{(S)-hydroxy[3-(4-hydroxyphenyl)propoxy]phosphoryl}adenosine | | Authors: | Hu, Y, Yin, L, Gai, Y, Wang, X.X, Wang, D.C. | | Deposit date: | 2010-06-14 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a Populus tomentosa 4-coumarate:CoA ligase shed light on its enzymatic mechanisms
Plant Cell, 22, 2010
|
|
6F7U
 
 | | Molecular Mechanism of ATP versus GTP Selectivity of Adenylate Kinase | | Descriptor: | Adenylate kinase, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Rogne, P, Rosselin, M, Grundstrom, C, Hedberg, C, Wolf-Watz, M, Sauer, U.H. | | Deposit date: | 2017-12-12 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular mechanism of ATP versus GTP selectivity of adenylate kinase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2A9Y
 
 | | Crystal structure of T. gondii adenosine kinase complexed with N6-dimethyladenosine | | Descriptor: | 6N-DIMETHYLADENOSINE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Zhang, Y, el Kouni, M.H, Ealick, S.E. | | Deposit date: | 2005-07-12 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Substrate analogs induce an intermediate conformational change in Toxoplasma gondii adenosine kinase
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2AA0
 
 | | Crystal structure of T. gondii adenosine kinase complexed with 6-methylmercaptopurine riboside | | Descriptor: | 2-HYDROXYMETHYL-5-(6-METHYLSULFANYL-PURIN-9-YL)-TETRAHYDRO-FURAN-3,4-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Zhang, Y, el Kouni, M.H, Ealick, S.E. | | Deposit date: | 2005-07-13 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate analogs induce an intermediate conformational change in Toxoplasma gondii adenosine kinase
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1DS5
 
 | | DIMERIC CRYSTAL STRUCTURE OF THE ALPHA SUBUNIT IN COMPLEX WITH TWO BETA PEPTIDES MIMICKING THE ARCHITECTURE OF THE TETRAMERIC PROTEIN KINASE CK2 HOLOENZYME. | | Descriptor: | ADENOSINE MONOPHOSPHATE, CASEIN KINASE, ALPHA CHAIN, ... | | Authors: | Battistutta, R, Sarno, S, De Moliner, E, Marin, O, Zanotti, G, Pinna, L.A. | | Deposit date: | 2000-01-07 | | Release date: | 2001-01-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | The crystal structure of the complex of Zea mays alpha subunit with a fragment of human beta subunit provides the clue to the architecture of protein kinase CK2 holoenzyme.
Eur.J.Biochem., 267, 2000
|
|
1J09
 
 | | Crystal structure of Thermus thermophilus glutamyl-tRNA synthetase complexed with ATP and Glu | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLUTAMIC ACID, Glutamyl-tRNA synthetase, ... | | Authors: | Sekine, S, Nureki, O, Dubois, D.Y, Bernier, S, Chenevert, R, Lapointe, J, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-12 | | Release date: | 2003-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding
EMBO J., 22, 2003
|
|
4ZME
 
 | |
4ZS4
 
 | |
5EJE
 
 | | Crystal structure of E. coli Adenylate kinase G56C/T163C double mutant in complex with Ap5a | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, COBALT (II) ION | | Authors: | Sauer, U.H, Kovermann, M, Grundstrom, C, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2015-11-01 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for ligand binding to an enzyme by a conformational selection pathway.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5IE3
 
 | | Crystal structure of a plant enzyme | | Descriptor: | ADENOSINE MONOPHOSPHATE, OXALIC ACID, Oxalate--CoA ligase | | Authors: | Fan, M.R, Li, M, Chang, W.R. | | Deposit date: | 2016-02-24 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Arabidopsis thaliana Oxalyl-CoA Synthetase Essential for Oxalate Degradation
Mol Plant, 9, 2016
|
|
4R1M
 
 | |
3SPD
 
 | | Crystal structure of aprataxin ortholog Hnt3 in complex with DNA | | Descriptor: | Aprataxin-like protein, DNA (5'-D(*GP*TP*CP*AP*CP*TP*AP*TP*CP*GP*GP*AP*AP*TP*GP*AP*G)-3'), DNA (5'-D(*TP*AP*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*AP*C)-3'), ... | | Authors: | Gong, Y, Zhu, D, Ding, J, Dou, C, Ren, X, Jiang, T, Wang, D. | | Deposit date: | 2011-07-01 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.912 Å) | | Cite: | Crystal structures of aprataxin ortholog Hnt3 reveal the mechanism for reversal of 5'-adenylated DNA
Nat.Struct.Mol.Biol., 18, 2011
|
|
