7R3H
 
 | |
8UW8
 
 | | Site-one protease and SPRING | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Membrane-bound transcription factor site-1 protease, ... | | Authors: | Kober, D.L. | | Deposit date: | 2023-11-06 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | SPRING licenses S1P-mediated cleavage of SREBP2 by displacing an inhibitory pro-domain.
Nat Commun, 15, 2024
|
|
1BBG
 
 | |
4ZL7
 
 | | Crystal structure of Pseudomonas aeruginosa DsbA E82I: Crystal I | | Descriptor: | HEXAETHYLENE GLYCOL, Thiol:disulfide interchange protein DsbA | | Authors: | McMahon, R.M, Martin, J.L. | | Deposit date: | 2015-05-01 | | Release date: | 2015-12-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Sent packing: protein engineering generates a new crystal form of Pseudomonas aeruginosa DsbA1 with increased catalytic surface accessibility.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
2MBS
 
 | | NMR solution structure of oxidized KpDsbA | | Descriptor: | Thiol:disulfide interchange protein | | Authors: | Kurth, F, Rimmer, K, Premkumar, L, Mohanty, B, Duprez, W, Halili, M.A, Shouldice, S.R, Heras, B, Fairlie, D.P, Scanlon, M.J, Martin, J.L. | | Deposit date: | 2013-08-03 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | Comparative Sequence, Structure and Redox Analyses of Klebsiella pneumoniae DsbA Show That Anti-Virulence Target DsbA Enzymes Fall into Distinct Classes.
Plos One, 8, 2013
|
|
4EDU
 
 | | The MBT repeats of human SCML2 in a complex with histone H2A peptide | | Descriptor: | Histone H2A.J peptide, Sex comb on midleg-like protein 2 | | Authors: | Nady, N, Amaya, M.F, Tempel, W, Ravichandran, M, Arrowsmith, C.H. | | Deposit date: | 2012-03-27 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Histone recognition by human malignant brain tumor domains.
J.Mol.Biol., 423, 2012
|
|
8IXQ
 
 | | Structure of glycosyltransferase LmbT in complex with GDP and ergothioneine | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Glycosyltransferase, trimethyl-[(2S)-1-oxidanyl-1-oxidanylidene-3-(2-sulfanylidene-1,3-dihydroimidazol-4-yl)propan-2-yl]azanium | | Authors: | Mori, T, Sun, X, Abe, I. | | Deposit date: | 2023-04-02 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Function Analysis of the S-Glycosylation Reaction in the Biosynthesis of Lincosamide Antibiotics.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8ILA
 
 | | Crystal structure of LmbT from Streptomyces lincolnensis NRRL ISP-5355 in complex with substrates | | Descriptor: | (2~{S})-3-[2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-[(1~{R},2~{R})-1-azanyl-2-oxidanyl-propyl]-3,4,5-tris(oxidanyl)oxan-2-yl]sulfanyl-1~{H}-imidazol-5-yl]-2-(trimethyl-$l^{4}-azanyl)propanoic acid, GUANOSINE-5'-DIPHOSPHATE, Glycosyltransferase | | Authors: | Dai, Y, Qiao, H, Xia, M, Fang, P, Liu, W. | | Deposit date: | 2023-03-03 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Basis of Low-Molecular-Weight Thiol Glycosylation in Lincomycin A Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
9NNR
 
 | |
9OA7
 
 | | GNAT family acetyltransferase EryM SeMet | | Descriptor: | GLYCEROL, Lysine N-acyltransferase MbtK, MAGNESIUM ION, ... | | Authors: | Li, Y, Smith, J. | | Deposit date: | 2025-04-19 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Redefining the role of the EryM acetyltransferase in natural product biosynthetic pathways.
Structure, 2025
|
|
9NNQ
 
 | | GNAT family acetyltransferase EryM | | Descriptor: | GLYCEROL, Lysine N-acyltransferase MbtK, MAGNESIUM ION, ... | | Authors: | Li, Y, Smith, J. | | Deposit date: | 2025-03-06 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Redefining the role of the EryM acetyltransferase in natural product biosynthetic pathways.
Structure, 2025
|
|
7DFM
 
 | |
7DFO
 
 | | Crystal structure of glycoside hydrolase family 11 beta-xylanase from Streptomyces olivaceoviridis E-86 in complex with 4-O-methyl-alpha-D-glucuronopyranosyl xylotetraose | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-[beta-D-xylopyranose-(1-4)]beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, CHLORIDE ION, ... | | Authors: | Fujimoto, Z, Kishine, N, Kaneko, S. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based substrate specificity analysis of GH11 xylanase from Streptomyces olivaceoviridis E-86.
Appl.Microbiol.Biotechnol., 105, 2021
|
|
6LUI
 
 | | Crystal structure of the SAMD1 WH domain and DNA complex | | Descriptor: | Atherin, DNA (5'-D(*AP*CP*CP*TP*GP*CP*GP*CP*AP*CP*CP*AP*T)-3'), DNA (5'-D(*AP*TP*GP*GP*TP*GP*CP*GP*CP*AP*GP*GP*T)-3') | | Authors: | Zhou, Y, Cao, Y, Wang, Z. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | The SAM domain-containing protein 1 (SAMD1) acts as a repressive chromatin regulator at unmethylated CpG islands.
Sci Adv, 7, 2021
|
|
5M7G
 
 | | Tubulin-MTD147 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(2,6-dimorpholin-4-ylpyridin-4-yl)-4-(trifluoromethyl)pyridin-2-amine, CALCIUM ION, ... | | Authors: | Bohnacker, T, Prota, A.E, Steinmetz, M.O, Wymann, M.P. | | Deposit date: | 2016-10-27 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.248 Å) | | Cite: | Deconvolution of Buparlisib's mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention.
Nat Commun, 8, 2017
|
|
1MBT
 
 | | OXIDOREDUCTASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, URIDINE DIPHOSPHO-N-ACETYLENOLPYRUVYLGLUCOSAMINE REDUCTASE | | Authors: | Benson, T.E, Walsh, C.T, Hogle, J.M. | | Deposit date: | 1995-11-28 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the substrate-free form of MurB, an essential enzyme for the synthesis of bacterial cell walls.
Structure, 4, 1996
|
|
3MBT
 
 | | Structure of monomeric Blc from E. coli | | Descriptor: | Outer membrane lipoprotein blc | | Authors: | Schiefner, A, Skerra, A. | | Deposit date: | 2010-03-26 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical analyses reveal a monomeric state of the bacterial lipocalin Blc.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2MBT
 
 | | NMR study of PaDsbA | | Descriptor: | Thiol:disulfide interchange protein DsbA | | Authors: | Rimmer, K, Mohanty, B, Scanlon, M.J. | | Deposit date: | 2013-08-03 | | Release date: | 2014-11-12 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Fragment library screening identifies hits that bind to the non-catalytic surface of Pseudomonas aeruginosa DsbA1.
PLoS ONE, 12, 2017
|
|
2YVE
 
 | | Crystal structure of the methylene blue-bound form of the multi-drug binding transcriptional repressor CgmR | | Descriptor: | 3,7-BIS(DIMETHYLAMINO)PHENOTHIAZIN-5-IUM, CHLORIDE ION, GLYCEROL, ... | | Authors: | Itou, H, Shirakihara, Y, Tanaka, I. | | Deposit date: | 2007-04-12 | | Release date: | 2008-04-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures of the Multidrug Binding Repressor Corynebacteriumglutamicum CgmR in Complex with Inducers and with an Operator
J.Mol.Biol., 403, 2010
|
|
3EAE
 
 | | PWWP domain of human hepatoma-derived growth factor 2 (HDGF2) | | Descriptor: | Hepatoma-derived growth factor-related protein 2 | | Authors: | Amaya, M.F, Zeng, H, Mackenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural and Histone Binding Ability Characterizations of Human PWWP Domains.
Plos One, 6, 2011
|
|
3LYI
 
 | | PWWP Domain of Human Bromodomain-Containing Protein 1 | | Descriptor: | Bromodomain-containing protein 1, CYSTEINESULFONIC ACID | | Authors: | Lam, R, Zeng, H, Ni, S, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-02-26 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and histone binding ability characterizations of human PWWP domains.
Plos One, 6, 2011
|
|
3MO8
 
 | | PWWP Domain of Human Bromodomain and PHD finger-containing protein 1 In Complex with Trimethylated H3K36 Peptide | | Descriptor: | Histone H3.2 TRIMETHYLATED H3K36 PEPTIDE, Peregrin | | Authors: | Lam, R, Zeng, H, Ni, S, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and Histone Binding Ability Characterizations of Human PWWP Domains.
Plos One, 6, 2011
|
|
3L42
 
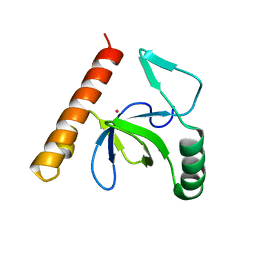 | | PWWP domain of human bromodomain and PHD finger containing protein 1 | | Descriptor: | Peregrin, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Zeng, H, Ni, S, Amaya, M.F, Dong, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural and Histone Binding Ability Characterizations of Human PWWP Domains.
Plos One, 6, 2011
|
|
3QBY
 
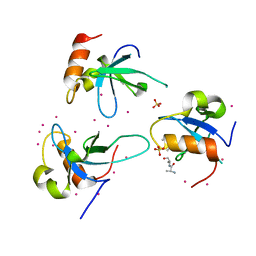 | | Crystal structure of the PWWP domain of human Hepatoma-derived growth factor 2 | | Descriptor: | H4K20me3 Histone H4 Peptide, Hepatoma-derived growth factor-related protein 2, SULFATE ION, ... | | Authors: | Zeng, H, Tempel, W, Amaya, M.F, Adams-Cioaba, M.A, Mackenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-14 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Histone Binding Ability Characterizations of Human PWWP Domains.
Plos One, 6, 2011
|
|
3PMI
 
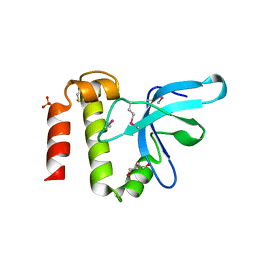 | | PWWP Domain of Human Mutated Melanoma-Associated Antigen 1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PWWP domain-containing protein MUM1, SULFATE ION, ... | | Authors: | Lam, R, Zeng, H, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-17 | | Release date: | 2010-12-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural and histone binding ability characterizations of human PWWP domains.
Plos One, 6, 2011
|
|
