4Q0A
 
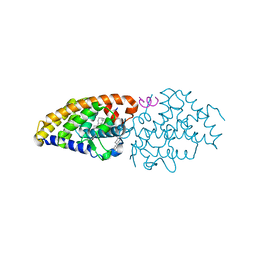 | | Vitamin D Receptor complex with lithocholic acid | | Descriptor: | (3beta,5beta,14beta,17alpha)-3-hydroxycholan-24-oic acid, Nuclear receptor coactivator 2, Vitamin D3 receptor A | | Authors: | Belorusova, A, Rochel, N. | | Deposit date: | 2014-04-01 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the molecular mechanism of vitamin d receptor activation by lithocholic Acid involving a new mode of ligand recognition.
J.Med.Chem., 57, 2014
|
|
4MNX
 
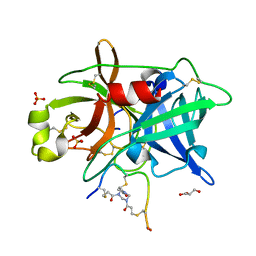 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK811 | | Descriptor: | 1,1',1''-(1,3,5-triazinane-1,3,5-triyl)tripropan-1-one, GLYCEROL, SULFATE ION, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2013-09-11 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Peptide ligands stabilized by small molecules.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4OB2
 
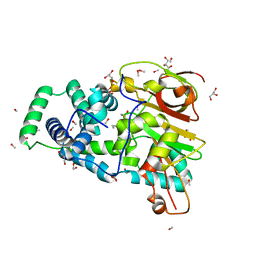 | | Crystal Structure of Nitrile Hydratase from Pseudonocardia thermophila bound to Butaneboronic Acid via Crystal Soaking | | Descriptor: | 1-BUTANE BORONIC ACID, COBALT (II) ION, Cobalt-containing nitrile hydratase subunit alpha, ... | | Authors: | Rui, W, Salette, M, Ruslan, S, Richard, H, Dali, L. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-26 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The active site sulfenic acid ligand in nitrile hydratases can function as a nucleophile.
J.Am.Chem.Soc., 136, 2014
|
|
4RPL
 
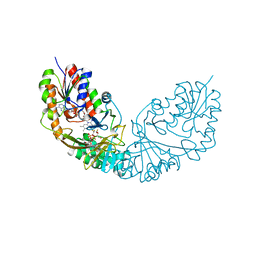 | | Crystal structure of Micobacterium tuberculosis UDP-Galactopyranose mutase in complex with tetrafluorinated substrate analog UDP-F4-Galp | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, UDP-galactopyranose mutase, [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,5S,6R)-3,3,4,4-tetrafluoro-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl dihydrogen diphosphate (non-preferred name) | | Authors: | Van Straaten, K.E, Sanders, D.A.R. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2499 Å) | | Cite: | Structural Basis of Ligand Binding to UDP-Galactopyranose Mutase from Mycobacterium tuberculosis Using Substrate and Tetrafluorinated Substrate Analogues.
J.Am.Chem.Soc., 137, 2015
|
|
4RPG
 
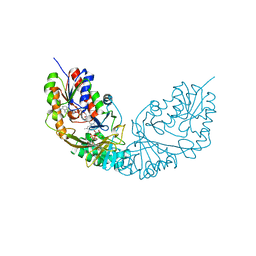 | |
1XCA
 
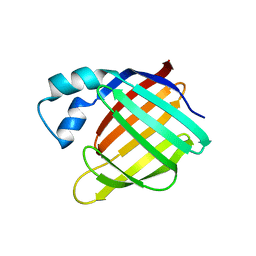 | | APO-CELLULAR RETINOIC ACID BINDING PROTEIN II | | Descriptor: | CELLULAR RETINOIC ACID BINDING PROTEIN TYPE II | | Authors: | Chen, X, Ji, X. | | Deposit date: | 1996-12-31 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of apo-cellular retinoic acid-binding protein type II (R111M) suggests a mechanism of ligand entry.
J.Mol.Biol., 278, 1998
|
|
4OB1
 
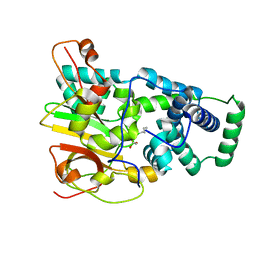 | | Crystal Structure of Nitrile Hydratase from Pseudonocardia thermophila bound to Butaneboronic Acid via Co-crystallization | | Descriptor: | 1-BUTANE BORONIC ACID, COBALT (II) ION, Cobalt-containing nitrile hydratase subunit alpha, ... | | Authors: | Rui, W, Salette, M, Ruslan, S, Richard, H, Dali, L. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | The active site sulfenic acid ligand in nitrile hydratases can function as a nucleophile.
J.Am.Chem.Soc., 136, 2014
|
|
6JXU
 
 | | SUMO1 bound to SLS4-SIM peptide from ICP0 | | Descriptor: | Small ubiquitin-related modifier, viral protein | | Authors: | Hembram, D.S.S, Negi, H, Shet, D, Das, R. | | Deposit date: | 2019-04-25 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Viral SUMO-Targeted Ubiquitin Ligase ICP0 is Phosphorylated and Activated by Host Kinase Chk2.
J.Mol.Biol., 432, 2020
|
|
3N5A
 
 | | Synaptotagmin-7, C2B-domain, calcium bound | | Descriptor: | CALCIUM ION, Synaptotagmin-7 | | Authors: | Tomchick, D.R, Rizo, J, Craig, T.K. | | Deposit date: | 2010-05-24 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.441 Å) | | Cite: | Structural and mutational analysis of functional differentiation between synaptotagmins-1 and -7.
Plos One, 5, 2010
|
|
1UMC
 
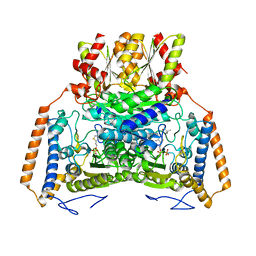 | | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 with 4-methylpentanoate | | Descriptor: | 2-oxo acid dehydrogenase alpha subunit, 2-oxo acid dehydrogenase beta subunit, 4-METHYL VALERIC ACID, ... | | Authors: | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-25 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ligand-induced Conformational Changes and a Reaction Intermediate in Branched-chain 2-Oxo Acid Dehydrogenase (E1) from Thermus thermophilus HB8, as Revealed by X-ray Crystallography
J.Mol.Biol., 337, 2004
|
|
2ARC
 
 | |
1YH2
 
 | | Ubiquitin-Conjugating Enzyme HSPC150 | | Descriptor: | HSPC150 protein similar to ubiquitin-conjugating enzyme | | Authors: | Walker, J.R, Avvakumov, G.V, Newman, E.M, Mackenzie, F, Kozieradzki, I, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
1STS
 
 | | STREPTAVIDIN DIMERIZED BY DISULFIDE-BONDED PEPTIDE FCHPQNT-NH2 DIMER | | Descriptor: | FCHPQNT-NH2, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T, Liu, B, Arze, R, Collins, N. | | Deposit date: | 1995-09-12 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Topochemical catalysis achieved by structure-based ligand design.
J.Biol.Chem., 270, 1995
|
|
1STR
 
 | | STREPTAVIDIN DIMERIZED BY DISULFIDE-BONDED PEPTIDE AC-CHPQNT-NH2 DIMER | | Descriptor: | AC-CHPQNT-NH2, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T, Liu, B, Arze, R, Collins, N. | | Deposit date: | 1995-09-12 | | Release date: | 1996-03-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Topochemical catalysis achieved by structure-based ligand design.
J.Biol.Chem., 270, 1995
|
|
2LAV
 
 | | NMR solution structure of human Vaccinia-Related Kinase 1 | | Descriptor: | Vaccinia-related kinase 1 | | Authors: | Shin, J, Yoon, H.S. | | Deposit date: | 2011-03-21 | | Release date: | 2011-05-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Human Vaccinia-related Kinase 1 (VRK1) Reveals the C-terminal Tail Essential for Its Structural Stability and Autocatalytic Activity.
J.Biol.Chem., 286, 2011
|
|
8X7I
 
 | | Cryo-EM structures of RNF168/UbcH5c-Ub in complex with H2AK13Ub nucleosomes determined by intein-based E2-Ub-NCP conjugation strategy | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Ai, H.S, Tong, Z.B, Deng, Z.H, Pan, M, Liu, L. | | Deposit date: | 2023-11-24 | | Release date: | 2024-08-07 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Capturing Snapshots of Nucleosomal H2A K13/K15 Ubiquitination Mediated by the Monomeric E3 Ligase RNF168
Biorxiv, 2024
|
|
8X7K
 
 | | Cryo-EM structures of RNF168/UbcH5c-Ub in complex with H2AK13Ub nucleosomes determined by activity-based chemical trapping strategy (adjacent H2AK13/15 dual-monoubiquitination) | | Descriptor: | DNA (143-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Ai, H.S, Tong, Z.B, Deng, Z.H, Pan, M, Liu, L. | | Deposit date: | 2023-11-24 | | Release date: | 2024-08-07 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Capturing Snapshots of Nucleosomal H2A K13/K15 Ubiquitination Mediated by the Monomeric E3 Ligase RNF168
Biorxiv, 2024
|
|
8X7J
 
 | | Cryo-EM structures of RNF168/UbcH5c-Ub/nucleosomes complex determined by activity-based chemical trapping strategy | | Descriptor: | DNA (144-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Ai, H.S, Tong, Z.B, Deng, Z.H, Pan, M, Liu, L. | | Deposit date: | 2023-11-24 | | Release date: | 2024-08-07 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Capturing Snapshots of Nucleosomal H2A K13/K15 Ubiquitination Mediated by the Monomeric E3 Ligase RNF168
Biorxiv, 2024
|
|
3OEA
 
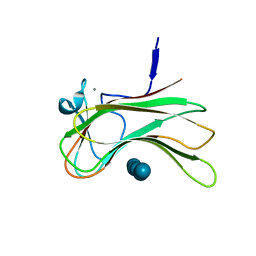 | | Crystal structure of the Q121E mutants of C.polysaccharolyticus CBM16-1 bound to cellopentaose | | Descriptor: | CALCIUM ION, S-layer associated multidomain endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Agarwal, V, Nair, S.K. | | Deposit date: | 2010-08-12 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Mutational insights into the roles of amino acid residues in ligand binding for two closely related family 16 carbohydrate binding modules.
J.Biol.Chem., 285, 2010
|
|
6UHN
 
 | | Crystal Structure of C148 mGFP-cDNA-1 | | Descriptor: | C148 mGFP-cDNA-1, UNKNOWN LIGAND | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
2OVQ
 
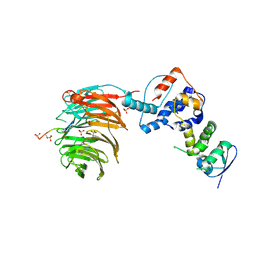 | | Structure of the Skp1-Fbw7-CyclinEdegC complex | | Descriptor: | F-box/WD repeat protein 7, S-phase kinase-associated protein 1A, SULFATE ION, ... | | Authors: | Hao, B, Oehlmann, S, Sowa, M.E, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2007-02-14 | | Release date: | 2007-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a Fbw7-Skp1-Cyclin E Complex: Multisite-Phosphorylated Substrate Recognition by SCF Ubiquitin Ligases
Mol.Cell, 26, 2007
|
|
4EH3
 
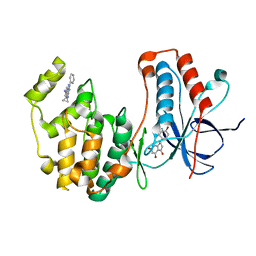 | | Human p38 MAP kinase in complex with NP-F2 and RL87 | | Descriptor: | Mitogen-activated protein kinase 14, NARINGENIN, N~4~-cyclopropyl-2-phenylquinazoline-4,7-diamine | | Authors: | Over, B, Gruetter, C, Waldmann, H, Rauh, D. | | Deposit date: | 2012-04-02 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Natural-product-derived fragments for fragment-based ligand discovery.
Nat Chem, 5, 2012
|
|
6UHQ
 
 | | Crystal Structure of C148 mGFP-cDNA-3 | | Descriptor: | C148 mGFP-cDNA-3, UNKNOWN LIGAND | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
2OVP
 
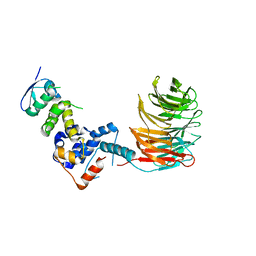 | | Structure of the Skp1-Fbw7 complex | | Descriptor: | F-box/WD repeat protein 7, S-phase kinase-associated protein 1A | | Authors: | Hao, B, Oehlmann, S, Sowa, M.E, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2007-02-14 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a Fbw7-Skp1-Cyclin E Complex: Multisite-Phosphorylated Substrate Recognition by SCF Ubiquitin Ligases
Mol.Cell, 26, 2007
|
|
8B0Z
 
 | |
