4KEL
 
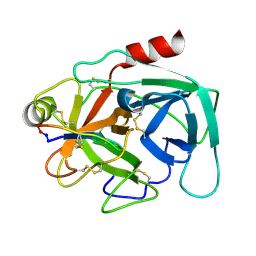 | | Atomic resolution crystal structure of Kallikrein-Related Peptidase 4 complexed with a modified SFTI inhibitor FCQR(N) | | Descriptor: | Kallikrein-4, Trypsin inhibitor 1 | | Authors: | Ilyichova, O.V, Swedberg, J.E, de Veer, S.J, Sit, K.C, Harris, J.M, Buckle, A.M. | | Deposit date: | 2013-04-25 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.148 Å) | | Cite: | KLK4 Inhibition by Cyclic and Acyclic Peptides: Structural and Dynamical Insights into Standard-Mechanism Protease Inhibitors.
Biochemistry, 58, 2019
|
|
7SUF
 
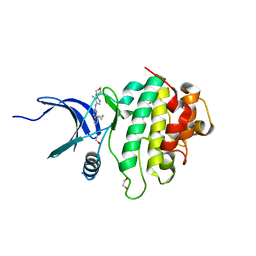 | | Structure of CHK1 10-pt. mutant complex with LRRK2 inhibitor 06 | | Descriptor: | 1,2-ETHANEDIOL, 8-cyclopropyl-N-[5-methyl-1-(oxan-4-yl)-1H-pyrazol-4-yl]quinazolin-2-amine, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure-Guided Discovery of Aminoquinazolines as Brain-Penetrant and Selective LRRK2 Inhibitors.
J.Med.Chem., 65, 2022
|
|
6Q2U
 
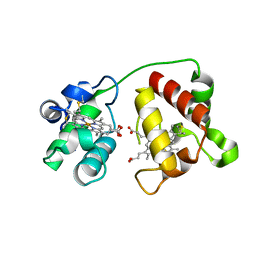 | | Structure of Cytochrome C4 from Pseudomonas aeruginosa | | Descriptor: | Cytochrome c4, HEME C | | Authors: | Carpenter, J.M, Zhong, F, Pletneva, E.V, Ragusa, M.J. | | Deposit date: | 2019-08-08 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and redox properties of the diheme electron carrier cytochrome c4from Pseudomonas aeruginosa.
J.Inorg.Biochem., 203, 2019
|
|
4KF4
 
 | | Crystal Structure of sfCherry | | Descriptor: | fluorescent protein sfCherry | | Authors: | Nguyen, H.B, Hung, L.-W, Yeates, T.O, Waldo, G.S, Terwilliger, T.C. | | Deposit date: | 2013-04-26 | | Release date: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Split green fluorescent protein as a modular binding partner for protein crystallization.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5I1B
 
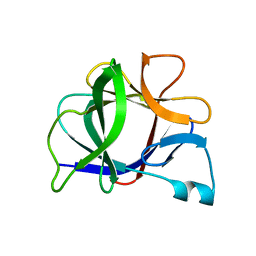 | |
7SUJ
 
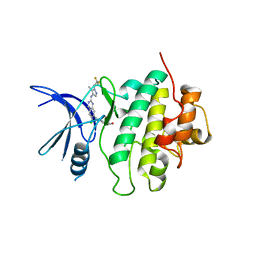 | | Structure of CHK1 10-pt. mutant complex with LRRK2 inhibitor 24 | | Descriptor: | (3R,4R)-4-{4-[6-chloro-2-({1-[(1R)-2,2-difluorocyclopropyl]-5-methyl-1H-pyrazol-4-yl}amino)quinazolin-7-yl]piperidin-1-yl}-4-methyloxolan-3-ol, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structure-Guided Discovery of Aminoquinazolines as Brain-Penetrant and Selective LRRK2 Inhibitors.
J.Med.Chem., 65, 2022
|
|
1SN4
 
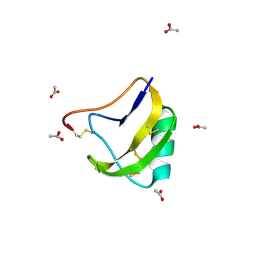 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M4 | | Descriptor: | ACETATE ION, PROTEIN (NEUROTOXIN BMK M4) | | Authors: | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | Deposit date: | 1998-11-11 | | Release date: | 1999-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
1JA4
 
 | |
7SVR
 
 | |
3M8K
 
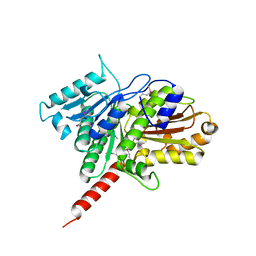 | | Protein structure of type III plasmid segregation TubZ | | Descriptor: | FtsZ/tubulin-related protein | | Authors: | Schumacher, M.A, Ni, L. | | Deposit date: | 2010-03-18 | | Release date: | 2010-07-07 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | From the Cover: Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1SN8
 
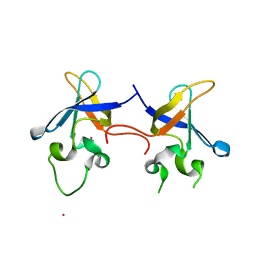 | | Crystal structure of the S1 domain of RNase E from E. coli (Pb derivative) | | Descriptor: | LEAD (II) ION, Ribonuclease E | | Authors: | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
1JAA
 
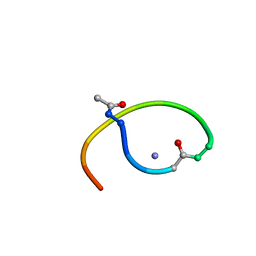 | | Solution structure of lactam analogue (DapE) of HIV gp41 600-612 loop. | | Descriptor: | DapE : (Ace)IWG(Dap)SGKLIETTA ANALOGUE OF HIV GP41 | | Authors: | Phan Chan Du, A, Limal, D, Semetey, V, Dali, H, Jolivet, M, Desgranges, C, Cung, M.T, Briand, J.P, Petit, M.C, Muller, S. | | Deposit date: | 2001-05-30 | | Release date: | 2003-07-01 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structural and immunological characterisation of heteroclitic peptide analogues corresponding to the 600-612 region of the HIV envelope gp41 glycoprotein.
J.Mol.Biol., 323, 2002
|
|
5I1G
 
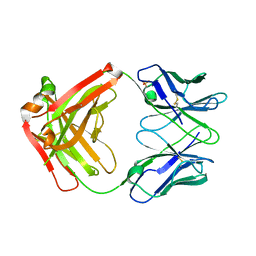 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV3-53/IGKV3-11 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, SULFATE ION | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2016-08-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
6PZJ
 
 | |
3DJ1
 
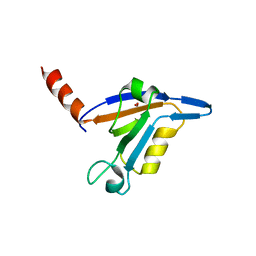 | | crystal structure of TIP-1 wild type | | Descriptor: | SULFATE ION, Tax1-binding protein 3 | | Authors: | Shen, Y. | | Deposit date: | 2008-06-21 | | Release date: | 2008-10-21 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of beta-Catenin Recognition by Tax-interacting Protein-1.
J.Mol.Biol., 384, 2008
|
|
1SNA
 
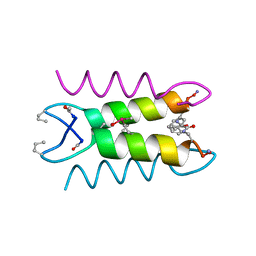 | | An Oligomeric Domain-Swapped Beta-Beta-Alpha Mini-Protein | | Descriptor: | ISOPROPYL ALCOHOL, tetrameric beta-beta-alpha mini-protein | | Authors: | Ali, M.H, Peisach, E, Allen, K.N, Imperiali, B. | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray structure analysis of a designed oligomeric miniprotein reveals a discrete quaternary architecture.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
6PZK
 
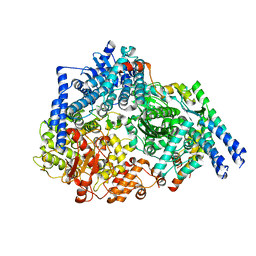 | |
1JAS
 
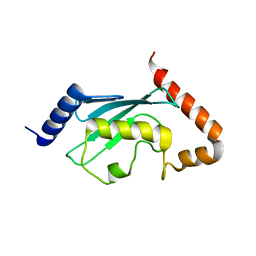 | | HsUbc2b | | Descriptor: | UBIQUITIN-CONJUGATING ENZYME E2-17 KDA | | Authors: | Miura, T, Klaus, W, Ross, A, Guentert, P, Senn, H. | | Deposit date: | 2001-05-31 | | Release date: | 2003-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the class I human ubiquitin-conjugating enzyme 2b
J.Biomol.NMR, 22, 2002
|
|
5I1Q
 
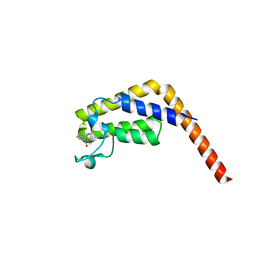 | | Second bromodomain of TAF1 bound to a pyrrolopyridone compound | | Descriptor: | 3-[6-(but-3-en-1-yl)-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl]-N,N-dimethylbenzamide, Transcription initiation factor TFIID subunit 1 | | Authors: | Tang, Y, Poy, F, Bellon, S.F. | | Deposit date: | 2016-01-09 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diving into the Water: Inducible Binding Conformations for BRD4, TAF1(2), BRD9, and CECR2 Bromodomains.
J.Med.Chem., 59, 2016
|
|
3M8V
 
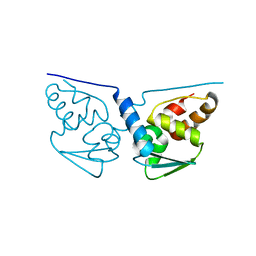 | |
1JC9
 
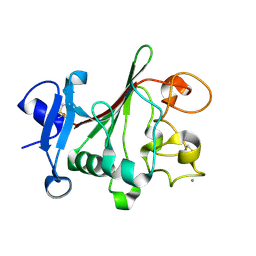 | | TACHYLECTIN 5A FROM TACHYPLEUS TRIDENTATUS (JAPANESE HORSESHOE CRAB) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, techylectin-5A | | Authors: | Kairies, N, Beisel, H.-G, Fuentes-Prior, P, Tsuda, R, Muta, T, Iwanaga, S, Bode, W, Huber, R, Kawabata, S. | | Deposit date: | 2001-06-08 | | Release date: | 2001-11-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The 2.0-A crystal structure of tachylectin 5A provides evidence for the common origin of the innate immunity and the blood coagulation systems.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
6Q07
 
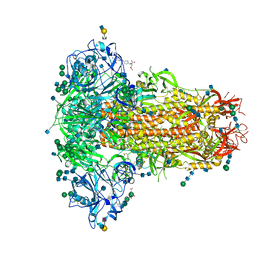 | | MERS-CoV S structure in complex with 2,6-sialyl-N-acetyl-lactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7SX4
 
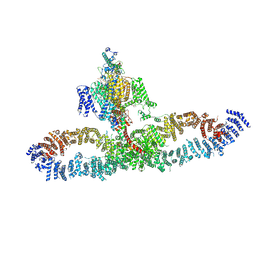 | | Human NALCN-FAM155A-UNC79-UNC80 channelosome with CaM bound, conformation 2/2 | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kschonsak, M, Chua, H.C, Weidling, C, Chakouri, N, Noland, C.L, Schott, K, Chang, T, Tam, C, Patel, N, Arthur, C.P, Leitner, A, Ben-Johny, M, Ciferri, C, Pless, S.A, Payandeh, J. | | Deposit date: | 2021-11-22 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural architecture of the human NALCN channelosome.
Nature, 603, 2022
|
|
4KHX
 
 | |
5I1Y
 
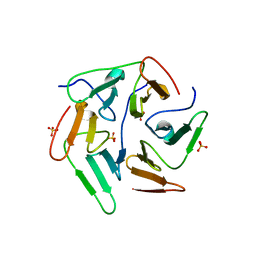 | | NvPizza2-H16S58 with cobalt | | Descriptor: | COBALT (II) ION, SULFATE ION, nvPizz2a-H16S58 | | Authors: | Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2016-02-07 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | NvPizza2-H16S58 with cobalt
To Be Published
|
|
